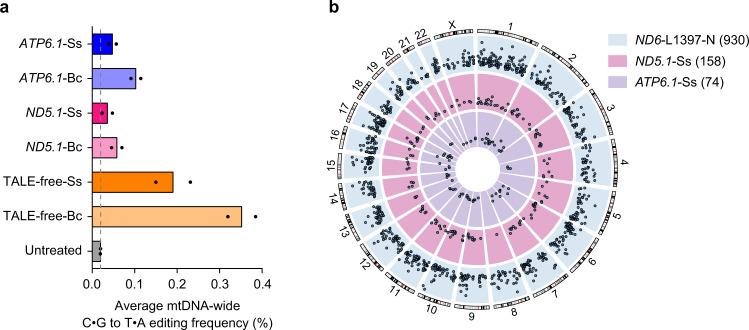
Fig. 4. Mitochondrial and nuclear genome-wide off-target editing by DdCBE_Ss.
a Average percentage of mtDNA-wide C•G to T•A off-target editing in untreated HEK293T cells and HEK293T cells treated with different DdCBEs. The vertical line represents the percentage of mtDNA C•G to T•A editing efficiency in untreated cells. Shown are the mean from n = 2 independent experiments. b Genome-wide circos plots representing the distribution and Detect-seq scores of identified nuclear DNA off-target sites on each chromosome for three different DdCBEs. The number of off-target sites is shown in parentheses. Source data are provided as a Source Data file.