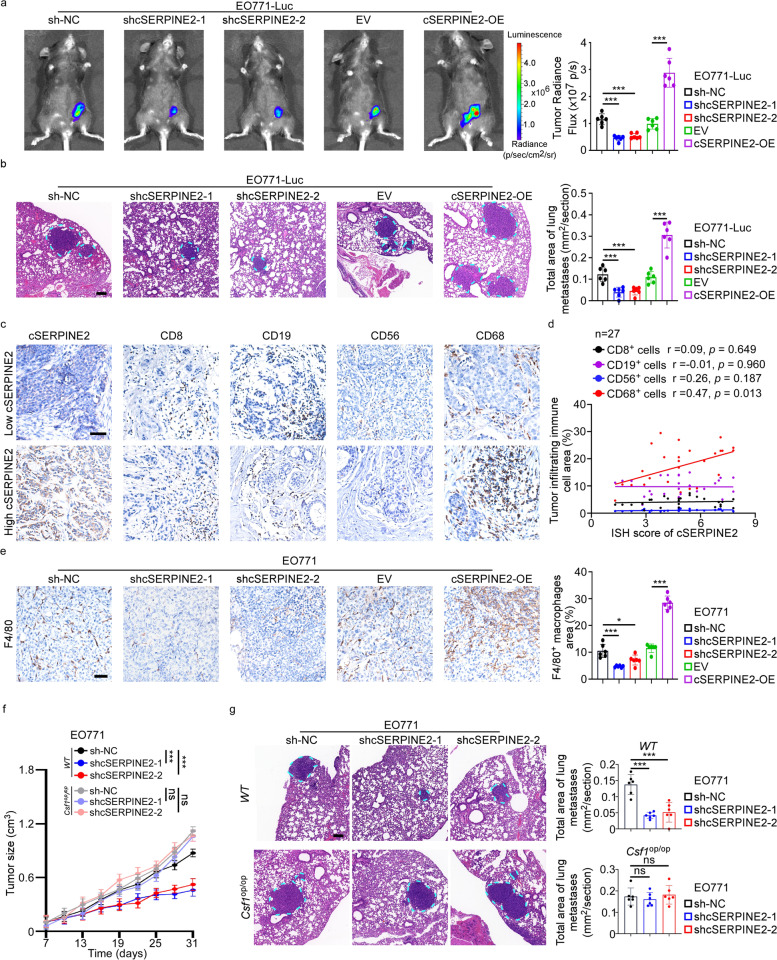
Fig. 2.
cSERPINE2 reshaped the immune microenvironment of breast cancer in mice. a Representative bioluminescence images (left) and quantification (right) of cSERPINE2 silenced and overexpressed tumor cells derived orthotopic breast cancer models (n = 6 mice for each group). b Representative lung H&E images (left) and quantification of lung metastatic area (right) of cSERPINE2 silenced and overexpressed tumor cells derived orthotopic breast cancer models (n = 6 mice for each group). Scale bar, 100 μm. c Representative IHC staining images of CD8+ T cells, CD19+ B cells, CD56+ NK cells and CD68+ macrophages in breast cancer tissues with low or high cSERPINE2 expression. Scale bar, 50 μm. d Correlation analysis of CD8+ T cells, CD19+ B cells, CD56+ NK cell and CD68+ macrophages infiltration and the cSERPINE2 expression in 27 breast cancer tissues. e Representative IHC staining images (left) and quantification (right) of F4/80+ macrophages infiltration in mice orthotopic breast cancer tissues. Scale bar, 50 μm. f, g cSERPINE2 silenced EO771 cells were used to constructed orthotopic tumor models in the wild-type (WT) or csf1op/op mice (n = 6 for each group). Tumor size was measured over time (f). Representative H&E images (left) and quantification (right) of lung metastases (g). Scale bar, 100 μm. Data are presented as the means ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001