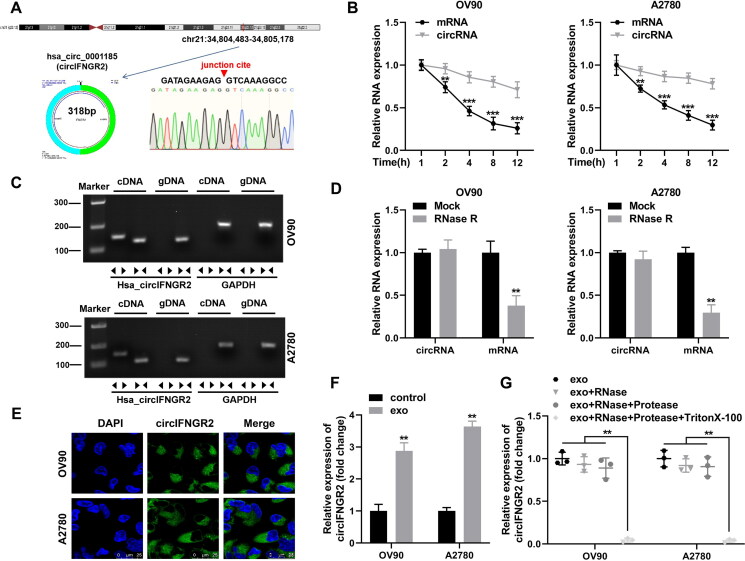
FIG 6.
CircIFNGR2 (hsa_circ_0001185) loop structure identification and cellular distribution. (A) Schematic diagram showing that circIFNGR2 originated from the IFNGR2 gene on chromosome 21q22.12, and Sanger sequencing proved the reverse splice junction sites of circIFNGR2. (B) qRT-PCR was applied to examine the transcript levels of linear IFNGR2 and circIFNGR2 in total RNA extracted from OV90 and A2780 cells processed with RNase R or mock, respectively. (C) Agarose gel electrophoresis was used to verify the presence of circIFNGR2 in A2780 and OV90 cells. (D) qRT-PCR was constructed to detect the stability of circIFNGR2 in OV90 and A2780 cells. (E) FISH was used to characterize the localization of circIFNGR2 in cells. (F) Relative expression levels of circIFNGR2 were measured by qRT-PCR in OVCA cells treated with or without exosomes. (G) Expression levels of circIFNGR2 in A2780 and OV90 cells treated with exo, exo + RNase, exo + RNase + Protease, exo + RNase + Protease + Triton X-100, respectively. **P < 0.01, ***P < 0.001.