Figure 1.
Identification of a Delta (AY.45)-Omicron (BA.1) recombinant virus with traces to New York and cytopathic effects
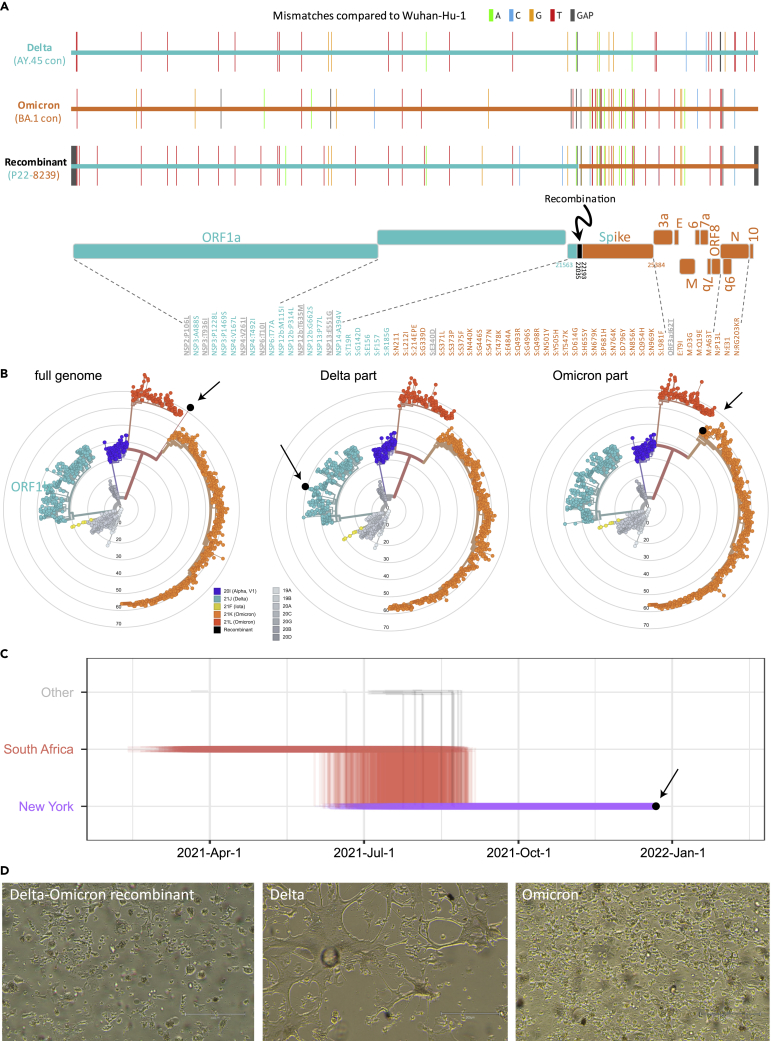
(A) Full genome mutations of the Delta-Omicron recombinant in comparison with Delta AY.45 and Omicron BA.1 consensus (con) sequences. In the schematic of the Delta and Omicron portions, all non-synonymous mutations are listed and shown in teal (Delta), orange (Omicron), or gray (not Delta or Omicron).
(B) Maximum likelihood phylogenies of 1557 global SARS-CoV-2 sequences based on a New York, New Jersey, and Connecticut-focused subsampling. The recombinant is shown in black. The phylogenetic analysis was done using the recombinant’s full genome (left) and Delta or Omicron parts only (middle and right). Concentrical rings indicate the number of mutations compared to Wuhan-Hu-1 (root).
(C) Markov jump trajectory plot depicting the ancestral location history of the AY.45 segment of the recombinant sequence. Each individual trajectory corresponds to the Markov jumps in a single tree from the posterior distribution of 1,800 trees (Figure S2B). Horizontal lines represent the time maintained at an ancestral location and vertical lines represent a Markov jump between two locations. The first SARS-CoV-2 positive test of the patient is indicated in black.
(D) Cytopathic effects observed in various lineages of SARS-CoV-2 on VeroE6/TMPRSS2 cells. Images were taken with an EVOS M5000 inverted microscope (ThermoFisher Scientific, Waltham, MA); 10X magnification. (see also Figures S1 and S2 and Tables S1 and S2).