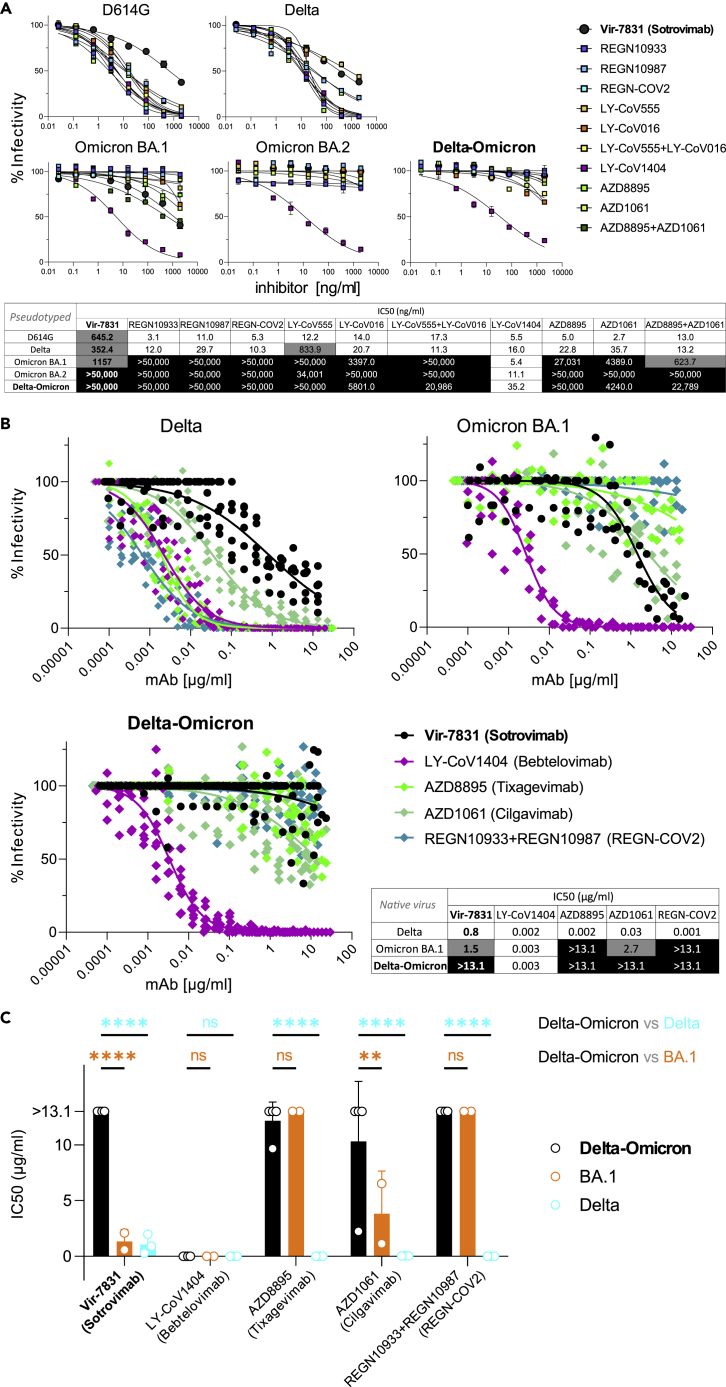
Figure 3.
Delta and Omicron (BA.1) are sensitive to Sotrovimab, but Delta-Omicron is resistant
(A) Neutralization of viruses with D614G, Delta, Omicron, and Delta-Omicron spike proteins by monoclonal antibodies (mAbs) Regeneron REGN10933 (Casirivamab), REGN10987 (Imdevimab), the REGN-COV-2 cocktail (Casirivimab + Imdevimab), LY-CoV555 (Bamlanivimab), LY-CoV016 (Etesevimab), the LY-CoV555+LY-CoV016 combination, LY-CoV1404 (Bebtelovimab), AZD8895 (Tixagevimab), AZD1061 (Cilgavimab), the AZD8895 + AZD1061 combination (Evusheld), and VIR-7831 (Sotrovimab). The heatmap table shows the half-maximal inhibitory concentrations (IC50) of the therapeutic mAbs calculated using the data from the antibody neutralization curves shown above. High resistance (IC50 > 3000 ng/mL) is highlighted in black, moderate resistance (50 ng/mL < IC50 < 3000 ng/mL) in gray, and no/low resistance (IC50 < 50 ng/mL) shown in white background.
(B) Neutralization of infectious native Delta-Omicron virus in comparison with Delta and BA.1 by a selection of five mAbs used in A, including Vir-7831 (Sotrovimab). Inhibition was determined in plaque reduction neutralization tests using serially diluted mAb doses. The graphs display all data points (staggered) from two to four biological replicates with technical duplicates (Delta-Omicron: four biological replicates). Neutralization curves are shown as non-linear regression fits for each mAb. In the IC50 heatmap table, high resistance (IC50 > 10 μg/mL) is highlighted in black, moderate resistance (1 μg/mL < IC50 < 10 μg/mL) in gray, and no/low resistance (IC50 < 1 μg/mL) shown in white background.
(C) Statistical comparison of IC50 values from all biological replicates in B (two-way ANOVA with Tukey’s multiple comparison test, ∗∗p < 0.005, ∗∗∗∗p < 0.0001, ns: not significant). Data in A and C are shown as mean with SD. (see also Table S3).