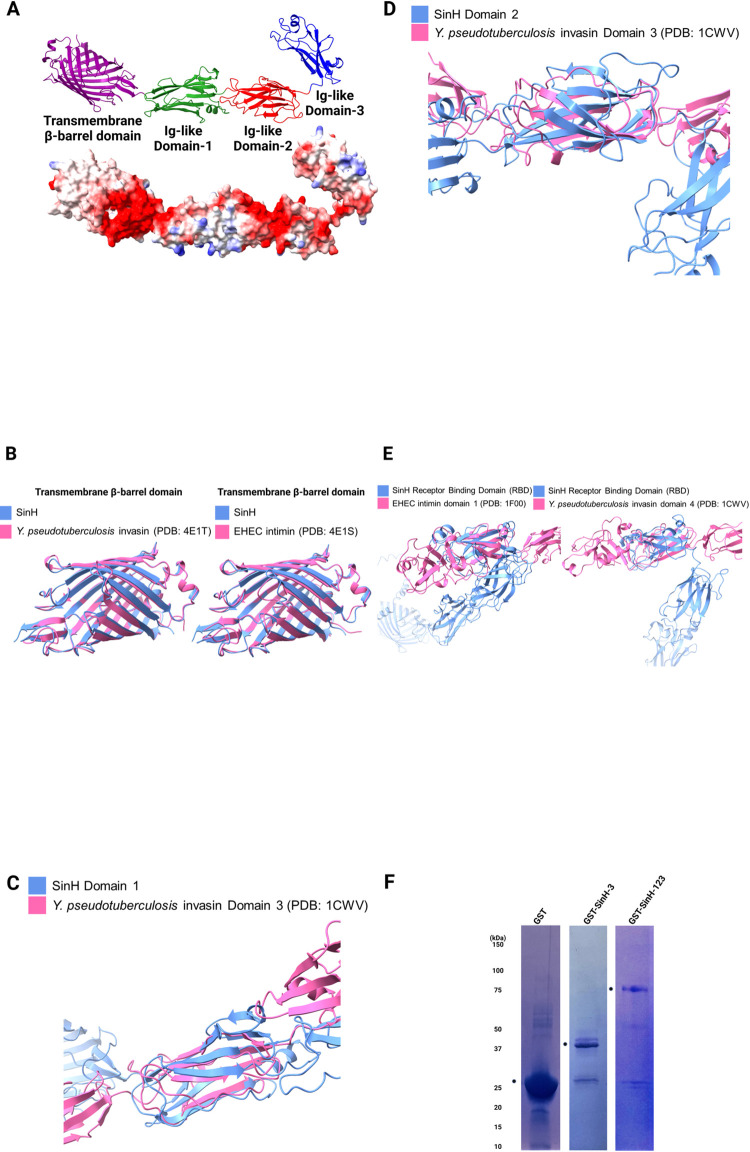
Fig 2. Structural alignment of predicted full-length SinH and expression and purification of SinH-based candidate antigens.
Structural alignments were generated by Pairwise Structure Alignment webserver, and aligned structures were visualized using ChimeraX and annotated with BioRender. (A) Predicted structure of full-length SinH protein (excluding disordered residues 1 through 101) with four distinct domains (Translocation β-barrel transmembrane domain: purple, Ig-like domain-1: green, Ig-like domain-2: red, Ig-like domain-3 (Receptor binding domain): blue). (B) Alignment between transmembrane β-barrel domains of predicted SinH protein structure (blue) and transmembrane domains of Y. pseudotuberculosis invasin (PDB: 4E1T) (red, left) and EHEC intimin (PDB: 4E1S) (red, right). (C) Alignment between domain-1 of SinH (blue) and domain-3 of Y. pseudotuberculosis invasin (red). (D) Alignment between domain-2 of SinH (blue) and domain-3 of Y. pseudotuberculosis invasin (red). (E) Alignment between the receptor-binding domain (RBD) of SinH (blue) and Ig-like domain-1 of EHEC intimin (red, left) and Ig-like domain-4 of Y. pseudotuberculosis invasin (red, right). (F) Genes encoding SinH-based antigens (Ig-like domain-1,2,3 or Ig-like domain-3) were cloned from ExPEC ST131 strain JJ1887. SinH-based antigens were recombinantly expressed with a glutathione-S-transferase (GST) tag and purified using immobilized GST-affinity chromatography. Purified antigens were separated and analyzed by SDS-PAGE and stained with Coomassie blue stain buffer. Predicted sizes of tagged proteins are as follows: GST-SinH-3, 40 kDa; GST-SinH-123, 70 kDa. Circle symbols indicate the locations of the GST-SinH Domain-3 and GST-SinH Domain-123, respectively, for each individual gel. The SDS-PAGE were annotated in BioRender.