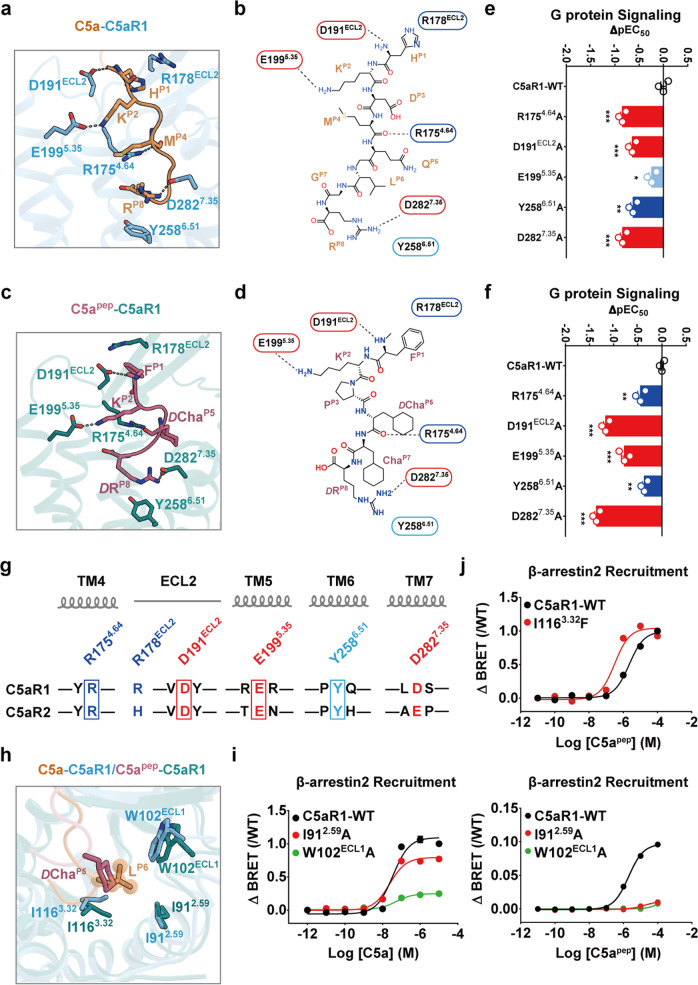
Fig. 3. Common and specific interactions between C5a and C5apep in site 2.
a–d Structural representation of common interaction sites between C5aR1 and C5a (a, b) or between C5aR1 and C5apep (c, d). The hook-shaped C-terminal tail of C5a or C5apep was anchored by polar or hydrophobic interactions with the residues R178ECL2, D191ECL2, E1995.35, R1754.64, D2827.35 and Y2586.51 in C5aR1. The detailed interactions between C5aR1 and C5a or C5apep are shown as 3D sticks (a, c) and 2D diagrams (b, d), respectively. Polar interactions are highlighted as black dashed lines. e, f The effects of mutations of common interaction sites on C5a (e) and C5apep (f) induced Gi protein signaling of C5aR1 examined by cAMP inhibition assay. Bars represent differences in calculated potency (ΔpEC50) for each mutant shown as percentage of the maximum in wild type. Data are the means ± SEM from at least three independent experiments, performed in triplicate and analyzed using one-way analysis of variance with Dunnett’s multiple comparison test to determine significance (compared with wild type). **P < 0.01, ***P < 0.001. g Sequence alignment of C5aR1 and C5aR2. The conserved common interaction sites are highlighted with squares. Residue positions labeled by Ballesteros–Weinstein numbering in C5aR1 are shown at the top. h Structural alignment of C5a-bound and C5apep-bound C5aR1 showing that Lp6 inserts into a hydrophobic cleft formed by I912.59, W102ECL1 and I1163.32. Instead, ChaP5 of C5apep cannot stretch out to the corresponding position of LP6 in C5a, leading to weaker interactions. i The I912.59A and W102ECL1A mutations greatly impaired C5apep-induced β-arrestin 2 recruitment compared with that of C5a; the signaling was monitored by BRET assay. Data are the means ± SEM from at least three independent experiments performed in triplicate. j The I1163.32F mutation increased the C5apep-induced β-arrestin 2 recruitment detected by BRET assay. Data are the means ± SEM from at least three independent experiments performed in triplicate.