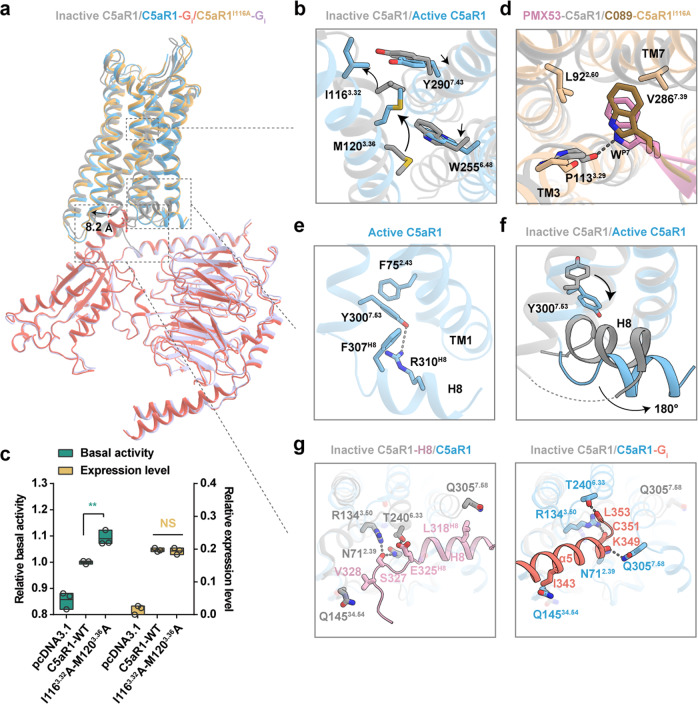
Fig. 5. The activation mechanism of C5aR1.
a Structural superposition of C5aR1–Gi complex (C5a-bound), C5aR1I116A–Gi complex (C089-bound) and inactive C5aR1 (PDB: 6C1R). b Structural comparison of the orthosteric binding pocket of inactive C5aR1 and active C5aR1. In particular, side chains of the residues I1163.32, M1203.36, W2556.48 and Y2907.43 are observed to exhibit a notable displacement in a “zipper-like” manner upon activation. The conformational changes of residues are shown as arrows. c The I1163.32A-M1203.36A mutation increases the basal activity of C5aR1. Statistical differences were determined by one-way ANOVA followed by the Dunnett’s multiple comparison test compared with C5aR1-WT. **P < 0.01; ns, no significance. Data are presented as the means ± SEM of three independent experiments performed in triplicate. d Structural alignment of PMX53–C5aR1 and C089–C5aR1I116A shows that WP7 in C089 adopts a similar pose to that in PMX53, inserting into the cleft between TM3 and TM7, contacting with L922.60, P1133.29 and V2867.39. e The non-canonical-state Y7.53 in active C5aR1 points toward TM1, and forms a hydrogen-bond with R310H8, as well as hydrophobic interactions with F752.43 and F307H8. f Conformational comparison of Y7.53 and H8 between inactive C5aR1 and active C5aR1. H8 swings away from the center of cytoplasmic surface with ~180° of flipping upon activation. g Key residues involved in stabilizing the H8 helix in the inactive state (PDB: 5O9H) (left panel) and participating in binding α5 helix of Gi protein in the active state (right panel).