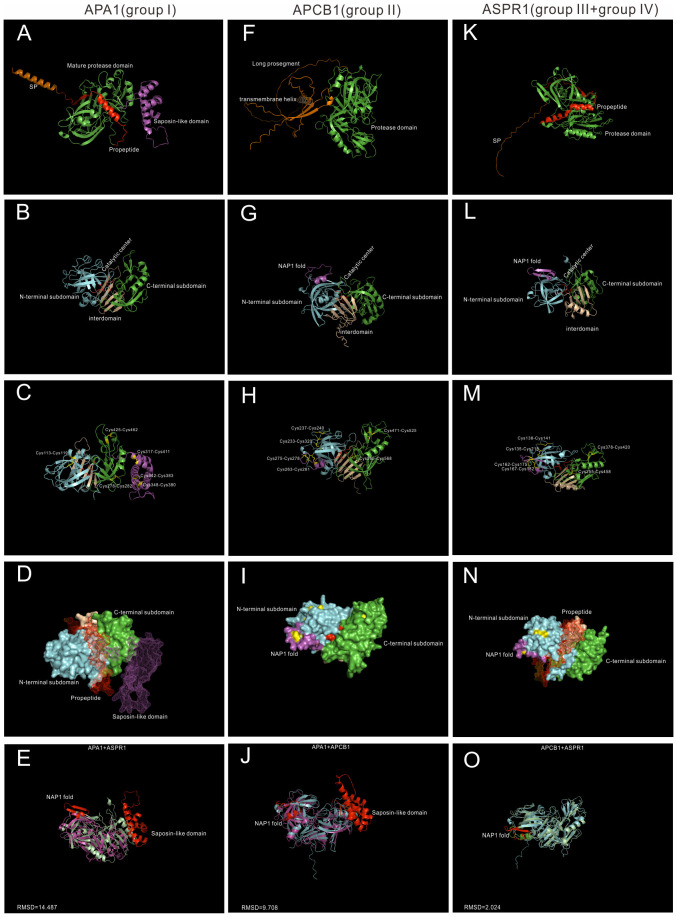
Figure 2.
AlphaFold predicted structure models of Group I-IV. (A–D) Group I typical structure (APA1). (A) overall structure of APA1. SP,signal peptide, is colored by orange; propeptide is colored by red, saposin-like domain is colored by purple. (B) Proposed mature protease domain of APA1. N-terminal subdomain is colored by cyan, C-terminal subdomain is colored by green, interdomain beta-sheet is colored by wheat, and active sites are colored by red. (C) Disulfide bonds of APA1. (D) Surface and mesh of APA. The proposed mature protease structure is presented by surface, propeptide and saposin-like domain is presented by mesh. (F–I) Group II typical structure (APCB1). (F) Overall structure of APCB1.Long prosegment is colored by orange and transmembrane helix is colored by wheat. (B) Proposed protease domain of APCB1,NAP1 fold is clored by purple. (H) Disulfide bonds of APCB1. (I) surface of APCB1. (K–N) Group III typical structure (ASPR1). (K) Overall structure of APSR1. (L) proposed mature protease structure of ASPR1. (M) Surface of ASPR1. (N) Disulfide bonds of ASPR1. (E–O) Superposition analysis of APA1, APCB1 and ASPR1, RMSD value are presented.