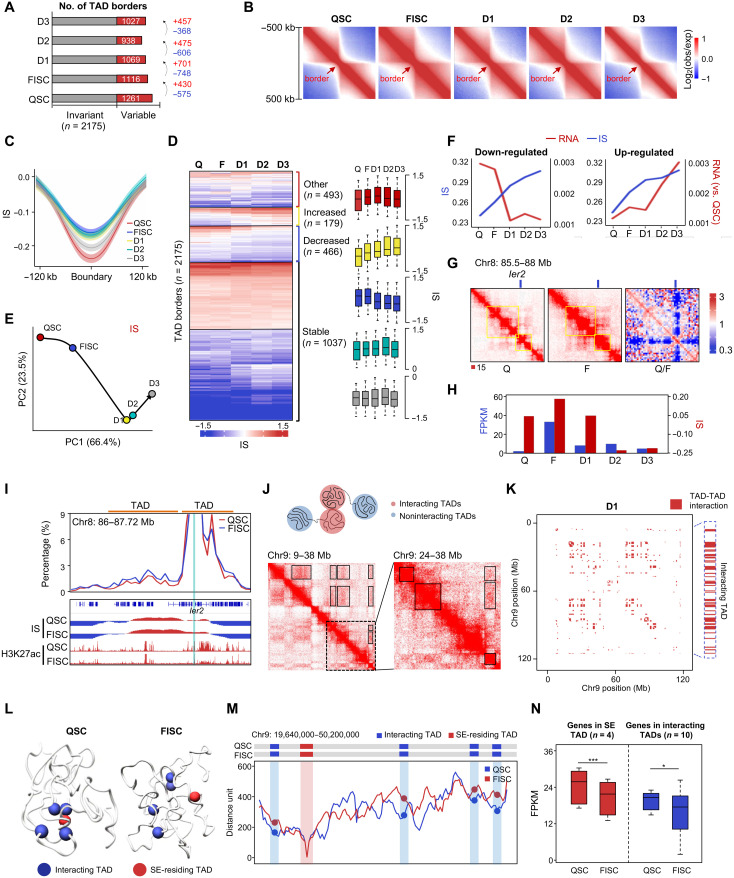
Fig. 2. Dynamic TAD organization during SC lineage development.
(A) Number of TAD borders. (B) Aggregate Hi-C maps centered on invariant TAD borders. Data are presented as the log ratio of observed and expected contacts in 40-kb bins. (C) Average IS in a 240-kb region centered on invariant TAD borders. Lines denote the mean values, while the shaded ribbons represent the 95% confidence interval. (D) k-means clustering (k = 20) of IS. Bar graphs show kinetics for groups that increased (n = 179), decreased (n = 466), transiently increased (n = 493), or did not change (n = 1037). (E) PCA of IS. Hypothetical trajectory is shown as black arrow. (F) Kinetics of expression changes at dynamic borders harboring up- or down-regulated genes with IS increased. (G) Illustration of in situ Hi-C contact maps (40-kb resolution) on the Ier2 locus. (H) IS kinetics of Ier2-residing TAD border and its expression dynamics. (I) Virtual 4C analysis on Ier2 locus. Ier2-residing and upstream TADs are indicated. Genome browser view of IS and H3K27ac ChIP-seq are shown. (J) Illustration of TAD-TAD interaction (top). A region of TAD-TAD interactions on chromosome 9 (D1) is visualized in the Hi-C data (bottom left). Boxes, TAD-TAD interactions. The dashed region is enlarged on the right. (K) Matrix of significant TAD-TAD interactions (red pixels) on chromosome 9 (D1); an example is highlighted (red pixels on the right side). (L) 3D chromatin conformation model of an SE-engaged TAD cluster. The SE-containing core TAD is marked in red, and other interacting TADs are in blue. (M) Line plot at 5-kb resolution displaying the median distance between SE-containing TAD and other interacting TADs. (N) Expression levels of genes (n = 4) in the above SE-containing TAD or in interacting TADs (n = 10). *P < 0.01 and ***P < 0.0001, Wilcoxon signed-rank test.