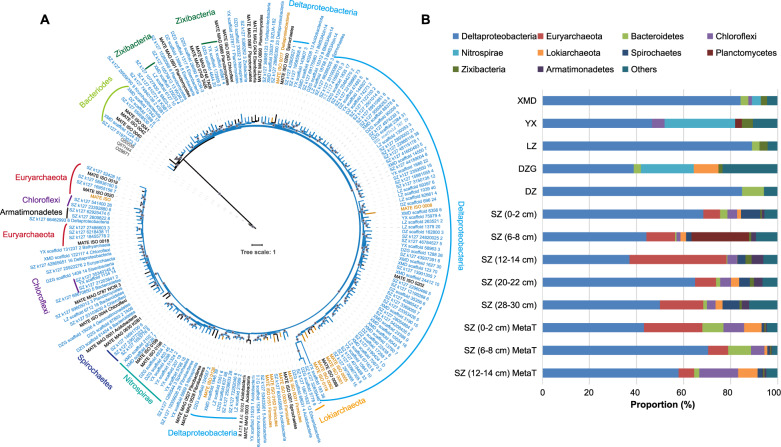
Fig. 2. Summary of 157 Metagenome-Assembled Genomes (MAGs) recovered with hgcA from mangrove sediments.
A Maximum-likelihood phylogenetic tree of HgcA amino acid sequences (1000 bootstraps; values >90% are shown by gray dots at the nodes). The 157 HgcA sequences from corresponded MAGs (Table S4) obtained in this study are highlighted in blue. HgcA sequences retrieved from public databases are shown in black. Experimentally confirmed HgcA are shown in brown. HgcA paralogues from non-methylating microorganisms were used as outgroups and are shown in gray. B Relative abundances of hgcA-carrying MAGs affiliated to different taxonomic groups in different mangrove sediment samples. Different taxonomic groups are represented by different colors. MetaT represent metatranscriptomic.