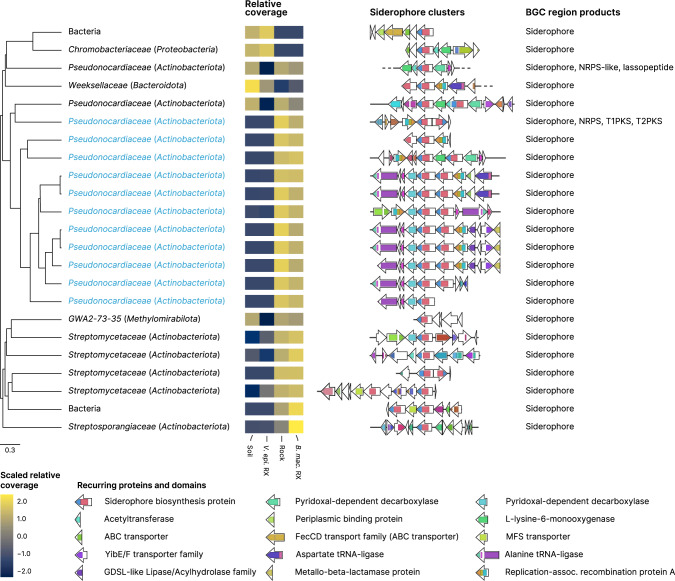
Fig. 6. Structural diversity of siderophore biosynthetic gene clusters (BGCs) identified in the campos rupestres metagenomes.
BGC regions containing siderophore clusters were hierarchically clustered using UPGMA with BiG-SCAPE distances. Groups of highly similar regions were identified based on their inconsistency coefficient and only the medoids are shown. Blue labels indicate the BGC regions that belong to the Pseudonocardiaceae-associated gene cluster clan. Taxonomies are presented at the family and phylum (in parenthesis) levels, except for two BGC regions whose contigs were assigned to the Bacteria domain. Heatmaps represent scaled means of log-transformed relative contig coverages in the four environments. Gene clusters are shown as arrays of genes (arrows) and their protein domains (colored blocks) centered at the siderophore biosynthesis protein. BGC regions containing other types of biosynthetic clusters (rightmost column) were trimmed to display only the loci assigned to the siderophore clusters.