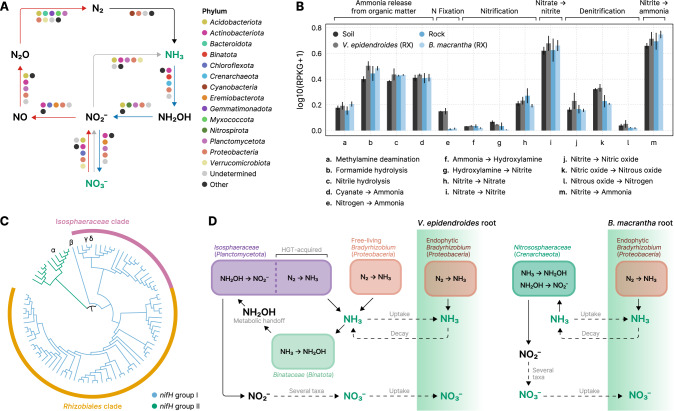
Fig. 7. The N-turnover landscape of the campos rupestres microbiomes.
A Phyla predicted to be involved in nitrogen-cycling reactions: fixation (black arrow), nitrification (blue arrows), denitrification (red arrows), and assimilatory nitrate reduction (gray arrow). Compounds that can be taken up by plants roots are depicted in green. Phyla that contributed less than 5% of the genes involved in each reaction were grouped under the “Other” category. B Total abundances (sum of RPKGs) of reactions involved in nitrogen turnover in the substrate and external root-associated communities. The abundances of multiprotein complexes (nifHDK, amoABC, nxrAB, narGHI, napAB, nasAB, norBC, nirBD, and nrfAH) were computed by averaging the abundances of their subunits. Vertical lines represent the standard error of the mean. RX = root (external). C Cladogram of phylogenies inferred from metagenomic nifH orthologs. Branches are colored according to the major group they belong to. The dominant clades are indicated by the outer rings. Orthologs encoded by MAGs encoding nif are indicated by greek letters: (α): Verrucomicrobiota, (β): Enterobacteriaceae, (γ and δ): Isosphaeraceae. D In the V. epidendroides-associated communities, nitrogen is mostly fixed by endophytic and free-living Bradyrhizobium, and by Isosphaeraceae, which likely received their nif complex via horizontal gene transfer (HGT). Ammonium is then oxidized into hydroxylamine by methylotrophic Binataceae. This molecule is then released from the cell and is oxidized into nitrite by Isosphaeraceae. In the B. macrantha-associated communities, nitrogen is likely converted into ammonium by endophytic Bradyrhizobium. Ammonia originated from organic matter decomposition is oxidized into hydroxylamine and, subsequently, into nitrite by Nitrososphaeraceae. In both plants, the oxidation of nitrite into nitrate is likely performed by a several taxa.