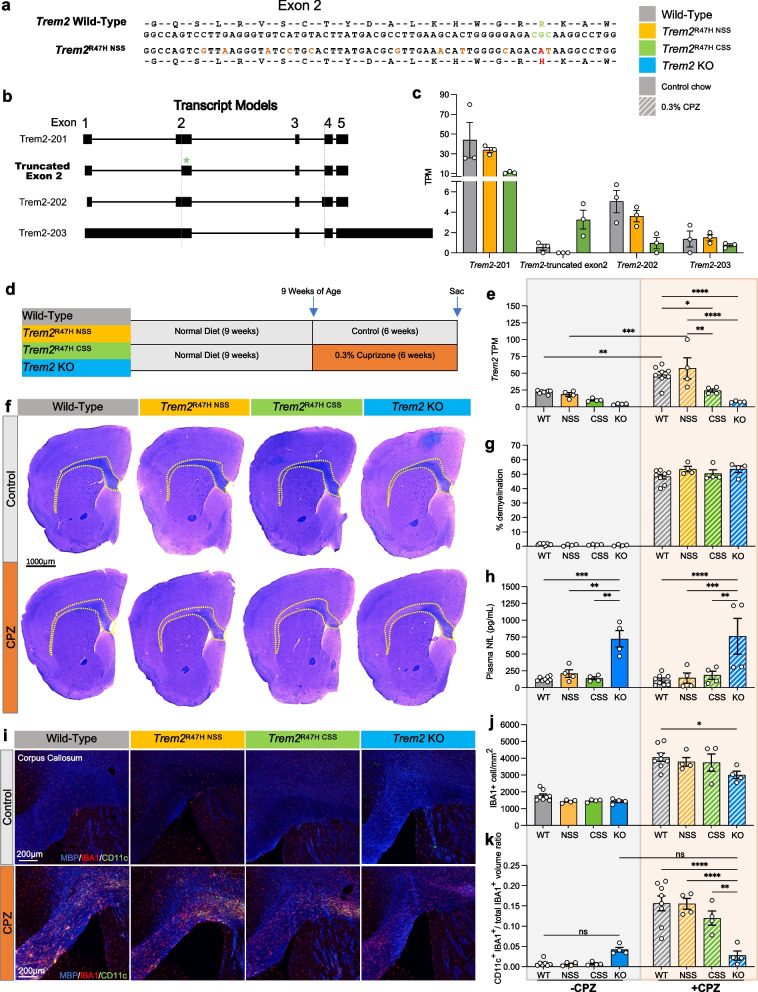
Fig. 1.
Cuprizone model of demyelination using wild-type, Trem2R47H NSS, Trem2R47H CSS, and Trem2 KO mice. a Amino acid coding sequence within exon 2 of mouse wild type Trem2 and Trem2 R47H NSS alleles. The triplet codon for arginine (R, green), the G to A transition that encodes histidine (H, red), and ten silent DNA mutation (tan) are shown in the Trem2R47H NSS allele. b Transcript models from long-read RNA-seq analysis showing Trem2 isoforms 201, 202, 203, and the truncated exon 2 from cryptic splicing reported in Trem2R47H CSS. The green asterisk denotes the truncated exon 2. c Trem2 isoform TPM values of wild-type, Trem2R47H NSS and Trem2R47H CSS. d Cuprizone (CPZ) feeding scheme of wild-type, Trem2R47H NSS, Trem2R47H CSS and Trem2 KO mice. e Trem2 TPM values of wild-type, Trem2R47H NSS, Trem2R47H CSS and Trem2 KO males examined from whole-brain bulk RNA-seq data showed similar increase in Trem2 expression in response to CPZ treatment in wild-type and Trem2R47H NSS that was attenuated in Trem2R47H CSS and Trem2 KO mice. f Representative whole-brain stitched images of wild-type, Trem2R47H NSS, Trem2R47H CSS and Trem2 KO males on control or CPZ diet stained for myelinated fibers with Luxol Fast Blue. Yellow dotted lines indicate region of demyelination. Scale bar = 1000 µm. g Quantification of demyelination reveals no change between genotypes. h Plasma neurofilament light-chain (NfL) quantified via Meso Scale Discovery suggests exacerbated axonal damage in Trem2 KO mice. i Representative images of corpus collosum from wild-type, Trem2R47H NSS, Trem2R47H CSS and Trem2 KO mice on control vs CPZ diet stained for myelin basic protein (blue), microglia (IBA1, red), and DAM gene marker (CD11c, green). j Quantification of IBA1+ cells per mm2 revealed expected increase in microgliosis in response to demyelination in CPZ-treated mice across all groups with Trem2 KO having fewer microglia than wild-type. k Quantification of CD11c+ microglia cell volume normalized to microglia volume. n = 4–8. Data are represented as mean ± SEM. Two-way ANOVA followed by Tukey’s post-hoc tests to examine biologically relevant interactions. Statistical significance is denoted by *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001