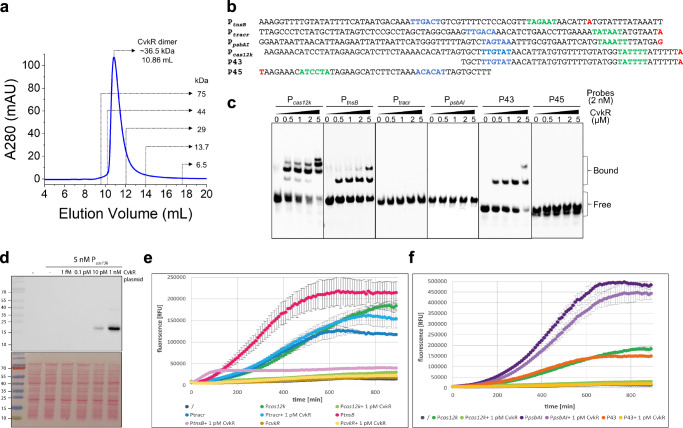
Fig. 5. Assays to test cas12k promoter elements.
a Molecular weight estimates of CvkR on size exclusion chromatography (SEC) with Superdex 75 10/300 GL column (GE Healthcare). Calibration standards are indicated. The elution volume of CvkR was ~10.86 mL, corresponding to a mass of ~36.5 kDa (twice its calculated molecular weight). mAU, milliabsorbance units. b Sequences of the tested promoter fragments. The probe P45 indicates the reverse complementary sequence of cvkR promoter. Putative −35 and −10 of cas12k, tnsB, tracrRNA, psbAI and cvkR promoter elements are shown in blue and green, respectively. The TSSs were included and marked in red. c EMSA assays showing the binding ability of CvkR to Pcas12k, PtnsB, Ptracr, PpsbAI and truncated fragments (P43 and P45) of Pcas12k. The data are presentative from two or three independent experiments. The free probe and complexes of protein-DNA complexes were marked as “Free” and “Bound”, respectively. d CvkR was expressed from vector pET28a and was detected by Western blot analysis via the N-terminal 6xHis tag (upper panel), and the stained membrane is shown in the lower panel (one out of three independent experiments is shown). The corresponding size for 6xHis-CvkR is 19.9 kDa. The prestained PageRuler (Thermo Fisher Scientific) was used as a size marker. e TXTL assay37 to compare the promoter activities of cas12k, cvkR, all3630 (tnsB) and tracrRNA (Pcas12k, PcvkR, PtnsB, and Ptracr) to drive deGFP expression in the absence or presence of CvkR. f The full-length version of Pcas12k and the P43 fragment encompassing 43 nt upstream of the cas12k TSS were tested in the TXTL system for their capacity to drive deGFP expression and mediate repression upon parallel expression of CvkR. PpsbAI was used as negative control for a promoter not controlled by CvkR. In both e and f, 1 pM CvkR plasmid was expressed together with the corresponding p70a plasmids (5 nM) with the respective promoter variants upstream of deGFP. Error bars show standard deviations calculated from two technical replicates in one representative example of three independent experiments. Source data are provided as a Source Data file.