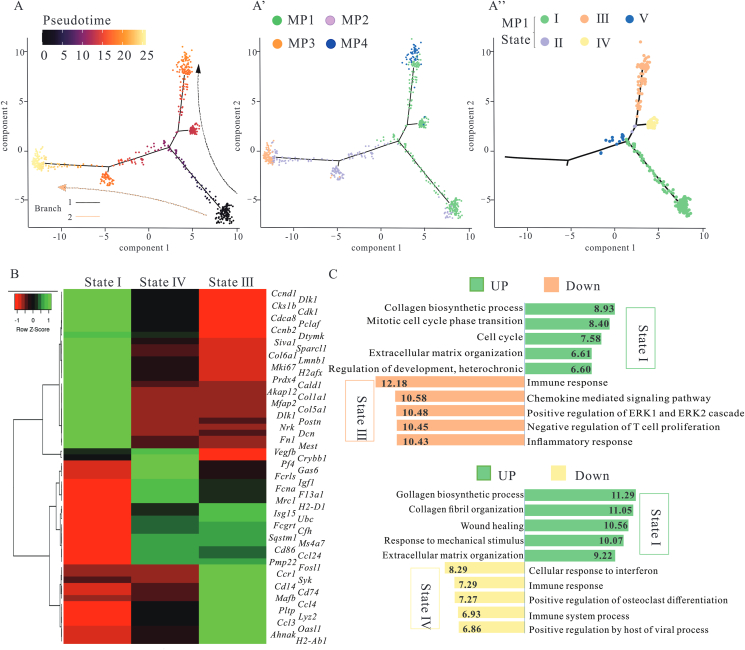
Figure 5.
MP1 cluster, a powerful and diverse function subset, had different derivations. (A) Ordering CRMs along a cell conversion trajectory using Monocle analysis. CRMs were color-labeled by pseudotime or sub-cluster. Cell clustering of MP1 based on the state along pseudotime trajectory showed five distinct trajectory states. (B) Heatmap of marker genes expression in the indicated states of MP1. (C) Selected top GO enrichment categories of the DEGs between different states. 30–50 cells per category were included.