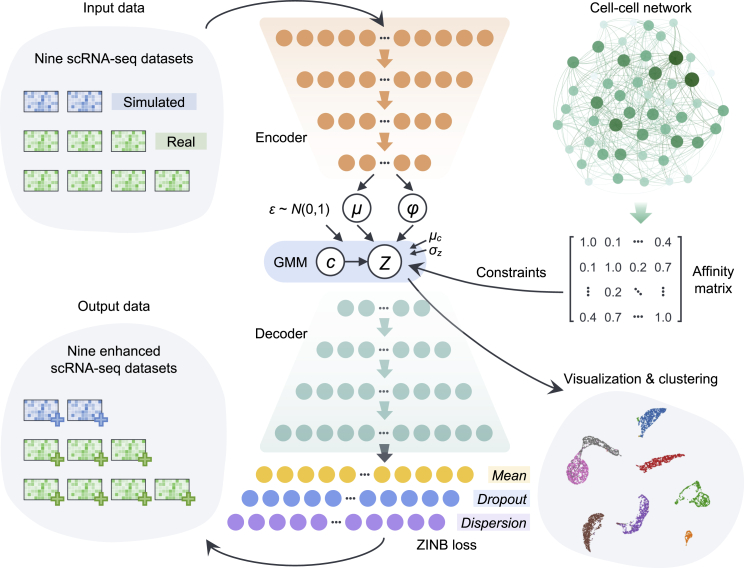
Figure 1.
Overview of the autoCell framework
autoCell uses a Gaussian mixture model (GMM) and a deep neural network (DNN) to model the process of data generation. It uses zero-inflated negative binomial (ZINB) loss to process “dropout” events in scRNA-seq. The encoder and decoder are a two-layer neural network (128–128) with 10-dimensional latent variables (features) directly connected to the output. The cell-cell network is used to constrain the latent feature ; thus, similar cells have similar latent features and cluster assignments. The yellow nodes depict the mean of the negative binomial distribution, which is the main output of the method representing denoised data, whereas the purple and blue nodes represent the other two parameters of the ZINB distribution, namely, dispersion and dropout.