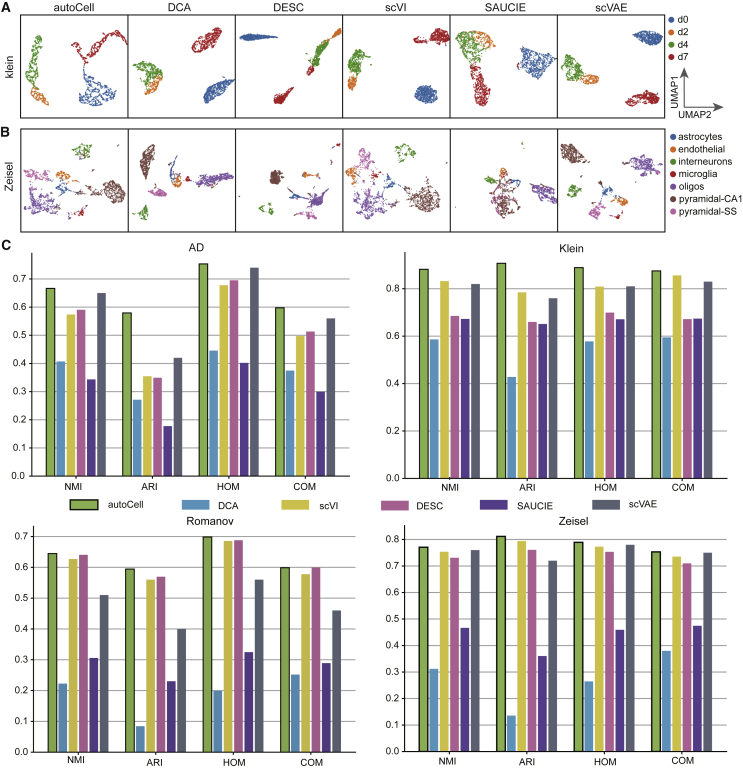
Figure 4.
UMAP visualization of the extracted features using different approaches
(A and B) We evaluated autoCell with DCA, DESC, scVI, SAUCIE, and scVAE using two datasets: (A) Klein and (B) Zeisel datasets. For comparison, autoCell, DCA, DESC, scVI, SAUCIE, and scVAE all performed dimension reduction to 10 dimensions before applying UMAP.
(C) Comparison on the effect of clustering on four benchmark datasets. Clustering accuracy was evaluated by applying K-means clustering on the extracted features to obtain cluster assignments.
NMI, normalized mutual information (the higher the value the better); ARI, adjusted rand index; COM, completeness (the higher the value the better); HOM, homogeneity (the higher the value the better).