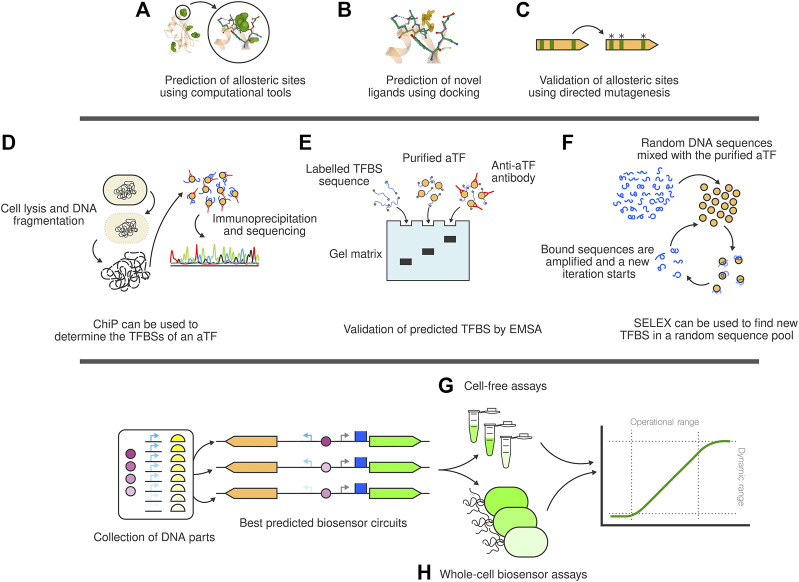
FIGURE 2.
The full stack biosensor development toolbox. (A) Computational tools can be used to determine the allosteric pockets of interaction between ligands and aTF. (B) Using these pockets as reference, docking computations can be carried out to assess the affinity of the aTF towards a library of putative ligand compounds. (C) The allosteric site computation can be validated using directed mutagenesis to evaluate changes in affinity or specificity. (D) ChIP technology allows researchers to determine the TFBSs of a newly discovered aTF. (E) Similarly, EMSA can be used to individually validate single TFBS. (F) SELEX can be used to artificially obtain new TFBS to the aTF. (G) Cell-free assays are a quick prototyping technology to test biosensor circuits once assembled. (H) Whole-cell biosensor experiments allow the characterisation of the biosensor circuit in conditions closer to in vivo applications.