Figure 2.
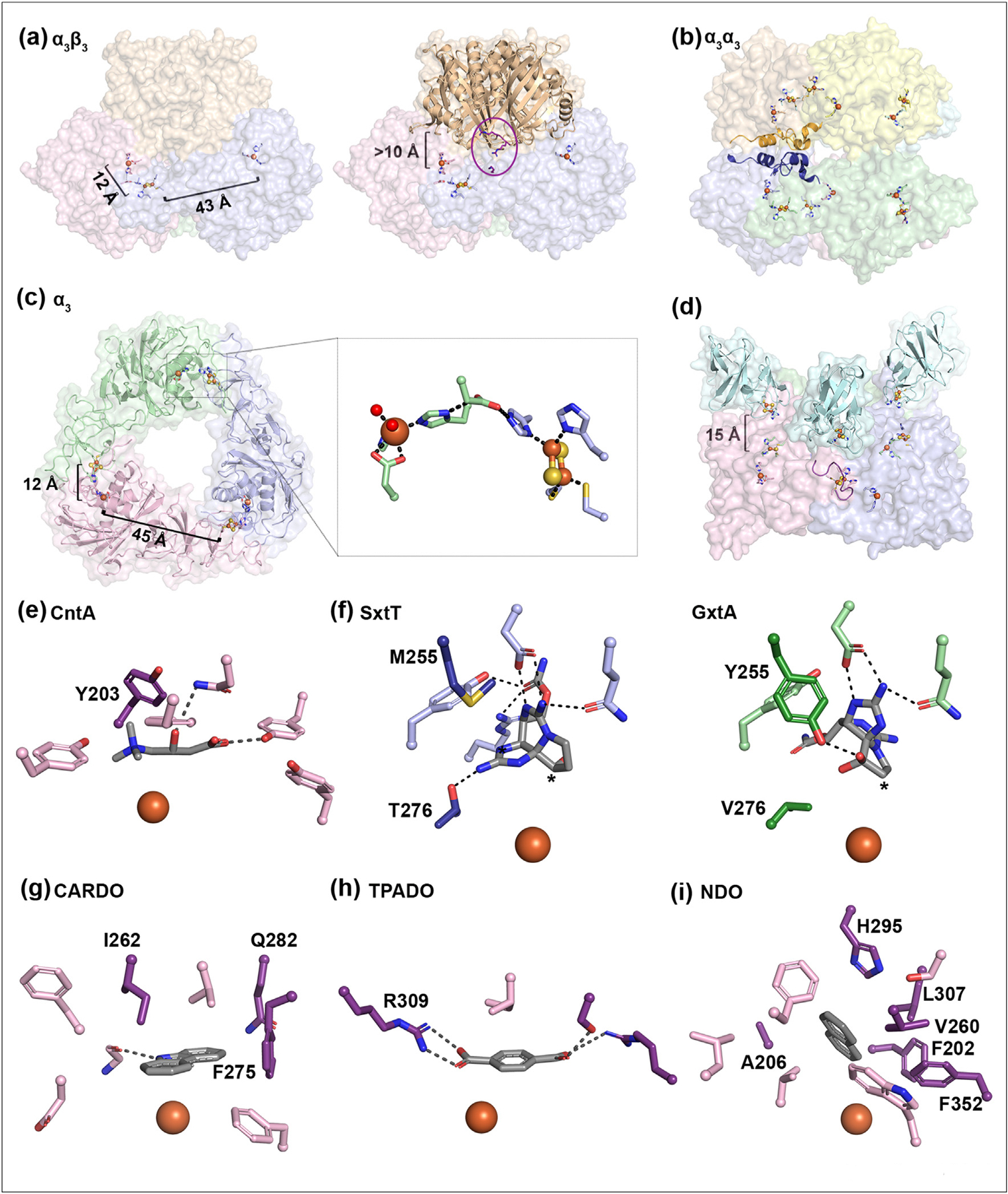
Rieske oxygenases are found in different architectures, each of which contains a trimer of α-subunits to coordinate the metallocenters and anchor the substrate near the mononuclear iron center. (a) In the α3β3 Rieske oxygenases, the metallocenters in a single α protomer are too far apart for electron transfer. However, assembly of the α subunits into a trimeric architecture moves the metallocenters from adjacent subunits close together (PDB:1NDO) [30]. The β subunits of the α3β3 enzymes sit more than 10 Å away from the active site. Residues implicated in binding the reductase are shown in an oval and highlighted in purple [49]. (b) The α3α3 architecture of PDO reveals that the trimers are staggered and likely stabilized by additional secondary structures found in each α subunit (dark blue and orange). (c) The α3 Rieske oxygenase architecture, as seen in the α3β3 and α3α3 architectures, arranges its Rieske cluster and mononuclear iron center across a subunit–subunit interface bridged by a conserved Asp residue (inset). (d) For the enzyme CARDO, three reductase proteins sit on top of the trimer such that the Fe–S clusters are aligned with the Rieske clusters of the oxygenase [43]. The loop that is suggested to stabilize the trimeric CARDO structure (and negate the need for a β subunit) is shown in purple for the light pink protomer. (e) The active site of CntA has a Tyr residue (Tyr203) that is important for substrate binding (PDB: 6Y9D). (f) Two active site residues in SxtT (left, PDB: 7SZH) and GxtA (right, PDB: 7SZE) (Met/Tyr255 and Thr/Val276) are important for positioning the native substrates for hydroxylation at the C12 and C11 positions, respectively (starred) [37,38]. Comparison between the panels reveals that the tricyclic substrate scaffold undergoes an approximate 120° rotation in the active site. (g) Residues Ile262, Gln282, and Phe275 are important to the selectivity of the CARDO-catalyzed reaction (PDB: 1WW9). (h) Arg309 in terephthalate dioxygenase (TPADO) is important for substrate binding and is conserved in additional Rieske oxygenases that perform chemistry on structurally similar molecules (PDB:7Q05). (i) Several residues (Phe202, Ala206, Val260, His295, Leu307, and Phe352) have been implicated in the selectivity of NDO-catalyzed reaction. In panels e, g, h, and i, specific residues involved in dictating reaction selectivity are shown in dark purple.
