Fig. 1 |. Definition of the human surfaceome on a genome-wide scale.
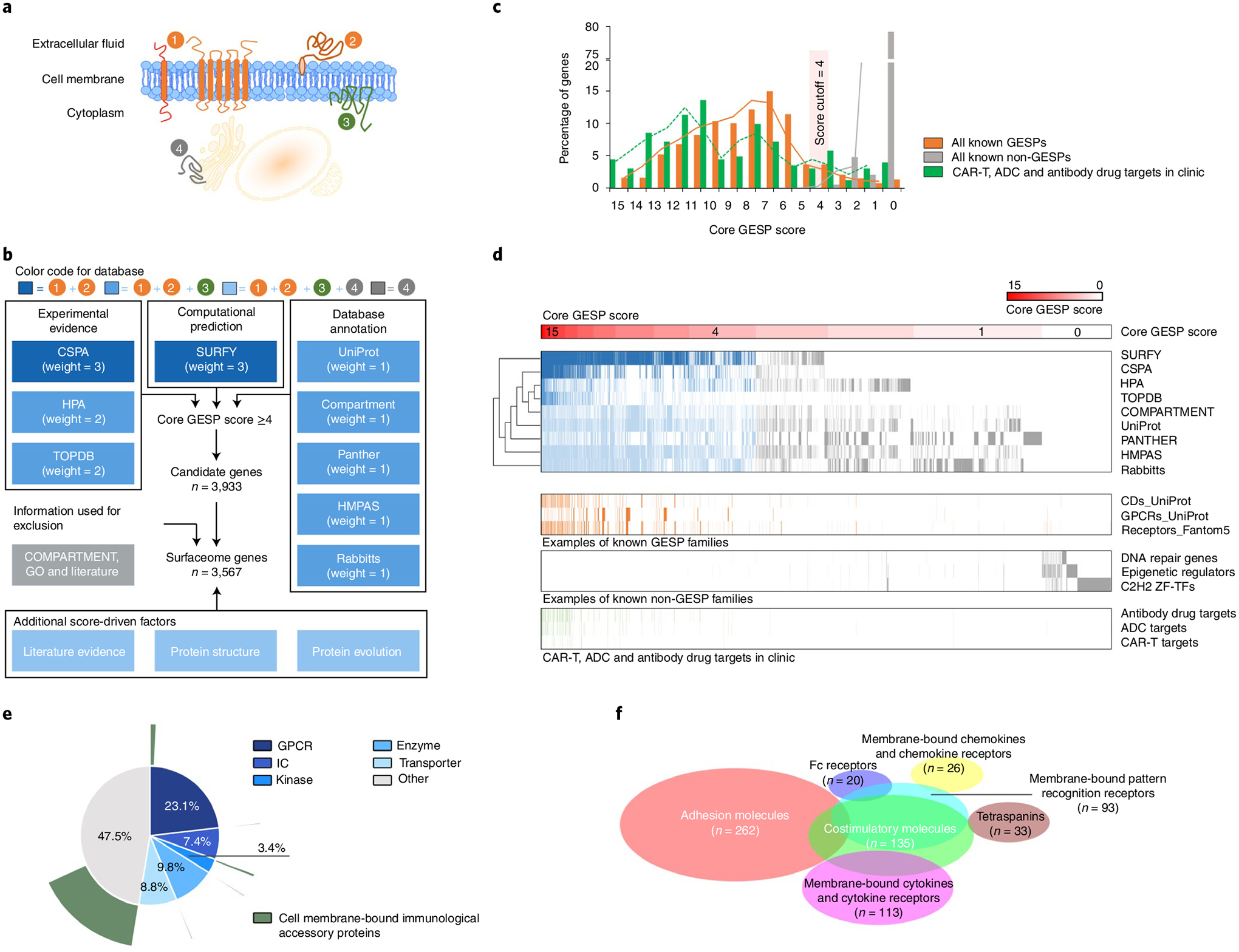
a, Schematic illustration of membrane proteins in cells. 1, Integral bitopic and polytopic proteins on the cell-surface membrane. 2, Integral monotopic proteins on the outer surface of the cell membrane. 3, Integral monotopic proteins on the inner surface of the cell membrane. 4, Membrane proteins on other intracellular membranes. b, The workflow to estimate final GESP scores for candidates of the GESPs. Nine independent and complementary resources were used to initially establish a core GESP score based on a weighted vote approach. Then COMPARTMENT, GO and literature searches were used to remove genes encoding proteins in intracellular membranes. Finally, other information and features were used as additional score-driven factors to estimate a final GESP score for each candidate. c, The distribution of the core GESP scores for known GESPs (orange) and non-GESPs (gray), as well as the target genes of CAR-Ts, ADCs and antibody drugs that are FDA approved or in clinical development (green). A core GESP score ≥4 was chosen as the cutoff to define GESPs. d, Heatmap showing genes across the nine surfaceome resources used in the present study, the examples of known GESPs and non-GESPs, and the target genes of CAR-Ts, ADCs and antibody drugs. Genes were ranked based on their core GESP scores. e, Classification of the GESPs based on gene superfamily category (inner circle), and the cell membrane-bound immunological accessory proteins highlighted in green (outer circle). f, Scaled Venn diagram showing the functional families among the cell mIAMs.
