Extended Data Fig. 1 |. Tissue specificity of GESPs across normal tissues.
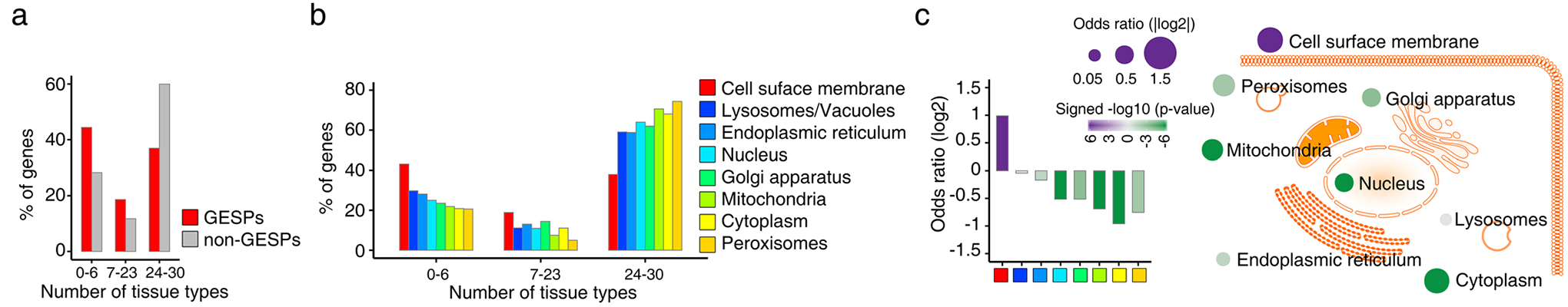
a, Percentages of genes which were detectable (median FPKM value >1) by RNA-seq analysis in 0–6, 7–23, and 24–30 tissue types. Red: GESPs; and gray: non-GESPs. b, The percentages of genes detectable in 0–6, 7–23, and 24–30 tissue types, stratified by subcellular location of gene products. c, Bar plot (left) and bubble plot (right) show enrichment of tissue type-specific genes in the corresponding subgroups based on subcellular location of gene products. P-values were calculated by two-sided Fisher’s exact test. Purple, enriched; green, depleted. The size of the bubble: absolute value of log2(odds ratio).
