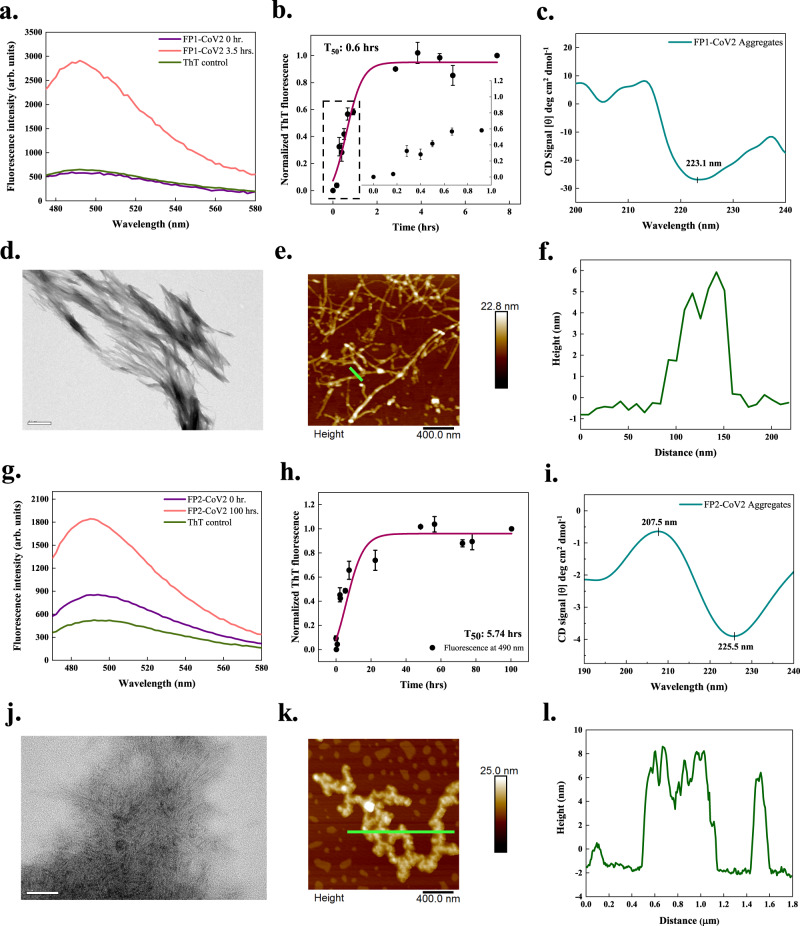
Fig. 4. In vitro aggregation of fusion peptides 1 (FP1-CoV2) and 2 (FP2-CoV2) of the SARS-CoV-2 S protein.
a, g ThT fluorescence scan (λex 440 nm) shows a ~6- and ~2-fold increment at λmax with FP1-CoV2 and FP2-CoV2 incubated samples, respectively. b, h Aggregation kinetics monitored using ThT fluorescence. Each dotted symbol represents the average value of two technical replicates and error bar represents the SEM of these replicates. The wine-coloured lines show the sigmoidal fittings of data points that determines the T50 values as 0.6 ± NC h and 5.74 ± NC h for FP1-CoV2 and FP2-CoV2 peptides, respectively. The kinetics reaction of FP1-CoV2 shows a very short lag phase that can be seen in the inset. c, i The secondary structure of 1 mg/ml peptides (both incubated for 240 h) investigated using CD spectroscopy shows a negative peak at ~223 and ~225 nm. The spectral data of FP1-CoV2 and FP2-CoV2 aggregates are smoothened with 4 and 10 points using the FFT filter function. d, j HR-TEM imaging of FP1-CoV2 (96 h) and FP2-CoV2 (100 h) exposed branched and long fibrils; scale bar represents 500 nm and 50 nm for images in panels (d) and (j). Sixteen and 22 TEM micrographs were captured for FP1-CoV2 and FP2-CoV2 peptides, respectively. e, k AFM images of FP1-CoV2 (96 h) and FP2-CoV2 (100 h) also reveal highly branched fibrils; scale bars represent 400 nm for both images. Seven and 5 AFM micrographs were captured for FP1-CoV2 and FP2-CoV2 aggregates, respectively. f, l Height profiles of FP1-CoV2 and FP2-CoV2 fibrils shown with green-coloured lines in panels (e) and (k), respectively. Having a height of ~8 nm, FP2-CoV2 amyloids are slightly bigger. Additional TEM and AFM micrographs are given in Supplementary Fig. 2. NC stands for not calculated. The source data are given in the Source Data file. Arb. units are arbitrary units.