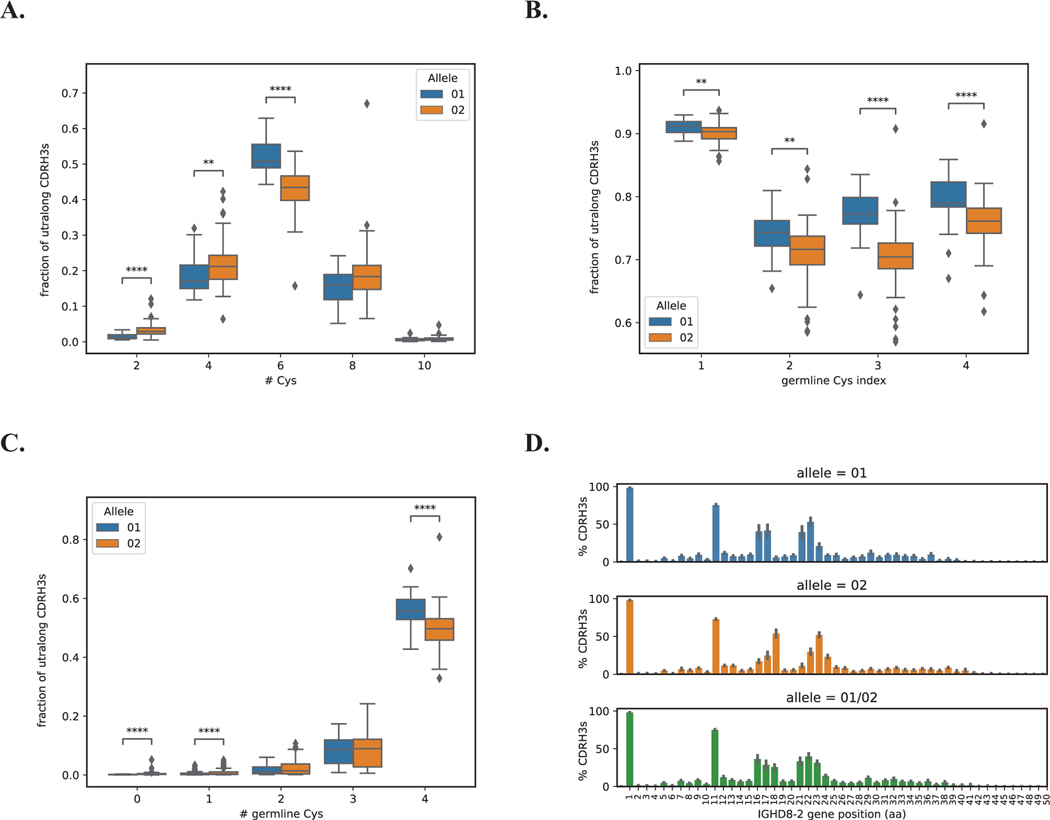
Figure 4. Allele usage and germline cysteine conservation and diversity in ultralong CDR H3 knobs.
Homozygous alleles IGHD8–2*01 and IGHD8–2*02 are shown in blue and orange, respectively. Heterozygous alleles (IGHD8–2*01/02) are shown in green. A. Fractions of ultralong CDRH3s with 2, 4, 6, 8, and 10 cysteines derived from homozygous alleles IGHD8–2*01 and IGHD8–2*02. B. Fractions of ultralong CDR H3s with conserved germline cysteines in homozygous alleles IGHD8–2*01 and IGHD8–2*02. C. Fractions of ultralong CDR H3s derived from homozygous alleles IGHD8–2*01 and IGHD8–2*02 with 0, 1, 2, 3, and 4 germline cysteines. D. Percentages of ultralong CDR H3 derived from the homozygous allele IGHD8–2*01 (top), the homozygous allele IGHD8–2*02 (middle), and heterozygous alleles IGHD8–2*01/02 (bottom) that have cysteines at positions corresponding to positions 1–50 of the germline D gene. CDR H3s were cropped and aligned by first position of the DH gene. P-values were computed using the Kruskal-Wallis test and denoted as follows: **<0.01, ****<0.0001. Non-significant P-values are not shown. Each box in Figure 4 shows the quartiles of the distribution. The whiskers show the rest of the distribution, except for outliers found using a function of the interquartile range implemented by the Seaborn package in Python.