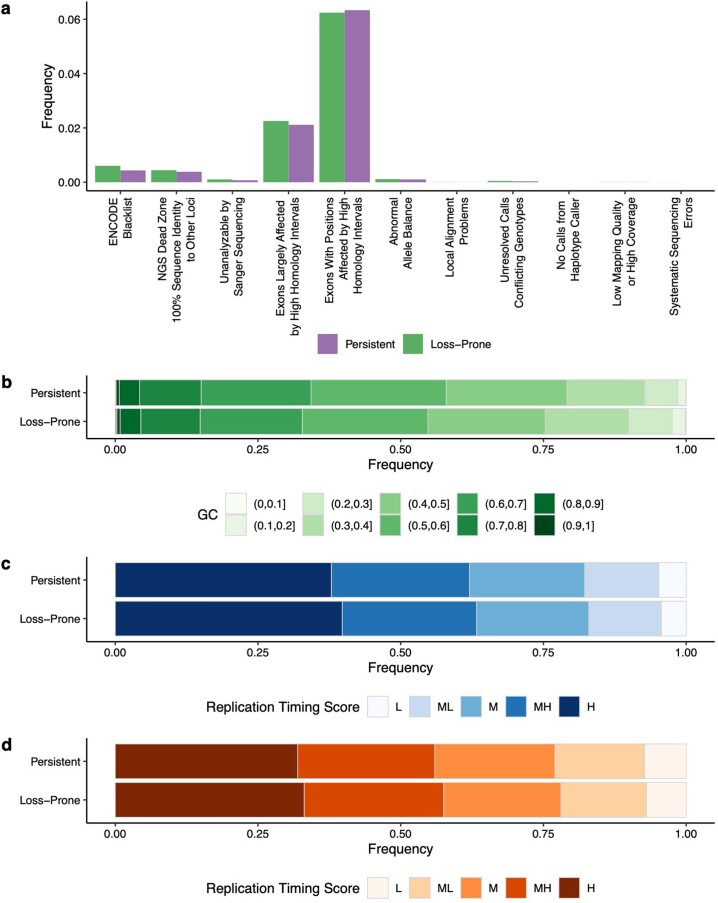
Extended Data Fig. 6. Assessment of genomic characteristics of loci harboring persistent vs loss-prone mutations in TCGA tumors.
(a) Genomic regions that are susceptible to limitations of NGS analysis, such as uncertain alignments or variant calls, were infrequent in the somatic mutation call set analyzed (MC3), and had similar representation in the persistent and loss-prone mutation categories (n = 9,242). (b) Similarly, the difference in GC composition of the immediate bases (9-mers) surrounding persistent and loss-prone mutations was negligible (n = 9,242; Cohen’s d = 0.08, persistent mean = 0.54, loss-prone mean = 0.52). Persistent and loss-prone mutations were found to have similar replication timing in (c) melanoma (TCGA-SKCM, n = 109, Cohen’s d = -0.035) and (d) NSCLC (combined set of TCGA-LUAD and TCGA-LUSC, n = 982, Cohen’s d = -0.032). Stacked bar plots depict the frequency of mutations in the five quantiles of replication timing. H, MH, M, ML, and L indicate mutations in the highest to lowest quintiles of replication timing in order; as an example mutations in earliest replicating regions are marked as H.