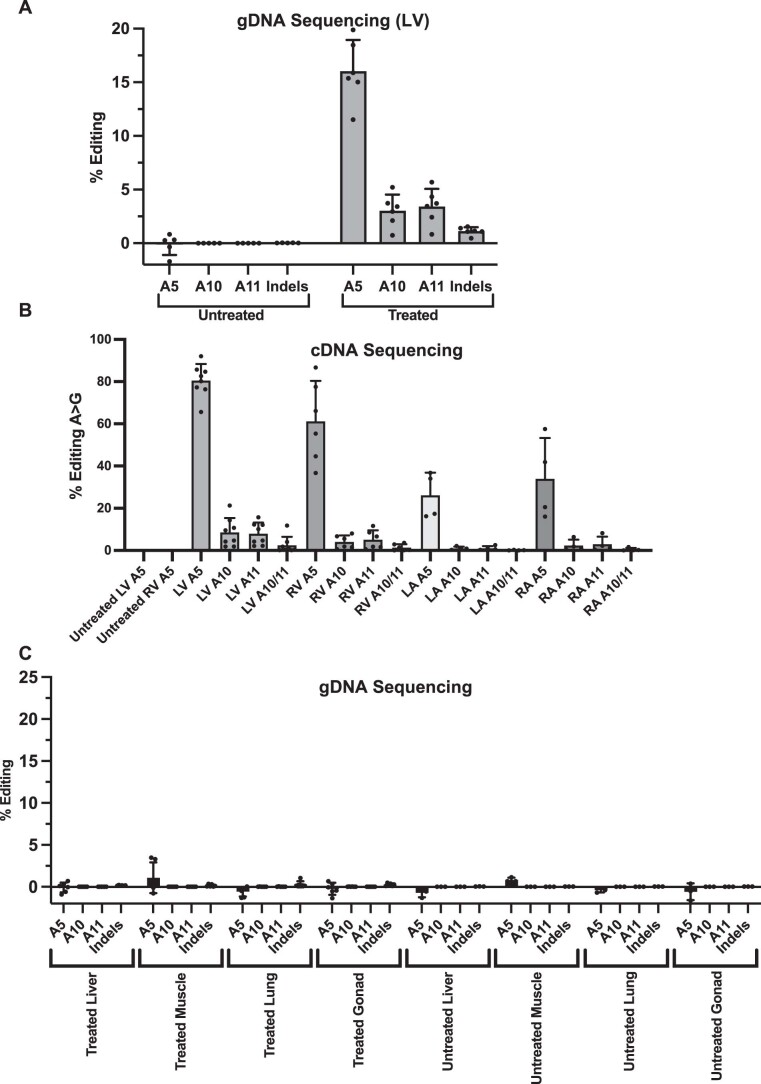
Extended Data Fig. 4. Bystander edits assessed after single injection of AAV9-ABE8e.
A) Editing efficiencies (%) measured after a single injection of AAV9-ABE8e based on high-throughput sequencing of PCR product amplified from LV gDNA (untreated n = 5, treated n = 6), quantified by CRISPResso2 (https://github.com/pinellolab/CRISPResso2). Editing efficiency at the on-target site A5 is compared to editing at bystander nucleotides A10 and A11. Indel rates were also assessed. B) Editing efficiencies (%) in untreated mice and mice treated with a single dose of AAV9-ABE8e. High-throughput sequencing of PCR product amplified from Myh6 cDNA derived from untreated LV and treated LV (n = 8), RV (n = 6), LA (n = 4) and RA (n = 4) identified bystander nucleotides A10, A11 or both (A10/11). C) Editing efficiency (%) assessed by sequencing PCR amplified gDNA extracted from non-targeted tissues: liver, skeletal muscle, lung and gonads from 6 treated and 3 untreated mice. Note low levels of on target Myh6 editing of the pathogenic variant R403Q (A5) in these non-targeted tissues. Data are presented as mean values ±SD.