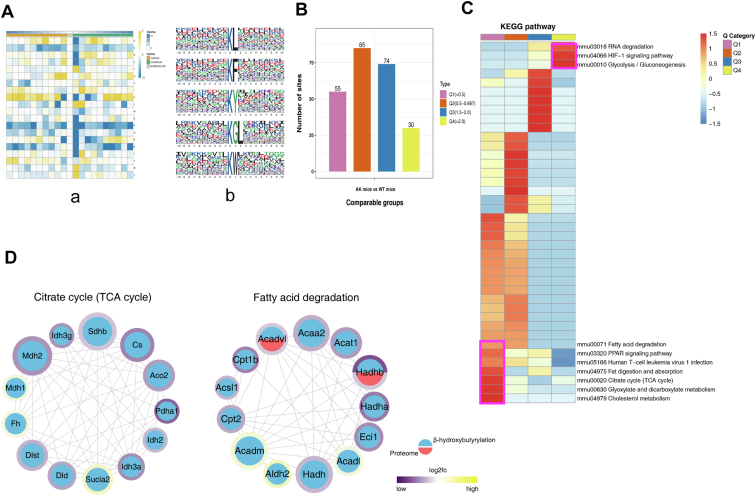
Fig. 6.
Characterizing Kbhb in mouse myocardium.A, the map shows the frequency change of amino acids close to the Kbhb site. Glycine (G), tryptophan (W), leucine (L), phenylalanine (F), and tyrosine (Y) were overexpressed, −1 and +1 positions around the Lys Kbhb sites. B and C, all Kbhb were equally divided into four groups according to their fold difference (Q1:< 0.5, Q2: 0.5–1/1.5, Q3: 1.5–2.0, Q4: >2.0). B, statistical chart of quantity of each group. C, KEGG pathway of the Kbhb proteome. Fat digestion and absorption, TCA cycle, cholesterol metabolism, and glyoxylate and dicarboxylate metabolism pathway were enriched with low AK/WT ratio. D, interaction network of differential Kbhb and proteome between TCA cycle and fatty acid degradation pathway. The Kbhb group is blue, while the protein group is red. The number of interacting nodes determines the size of the circle. In the outer circle, the ratio value following log2 treatment was displayed using the Omics Visualizer 1.3.0 plug-in. The color is more yellow the higher the value and more purple the lower the value. The protein’s location with the biggest difference served as the ratio value for the modified group (ranked by the absolute value of ratio after log2 treatment). Kbhb, Lysine β-hydroxybutyrylation; KEGG, Kyoto Encyclopedia of Genes and Genomes.