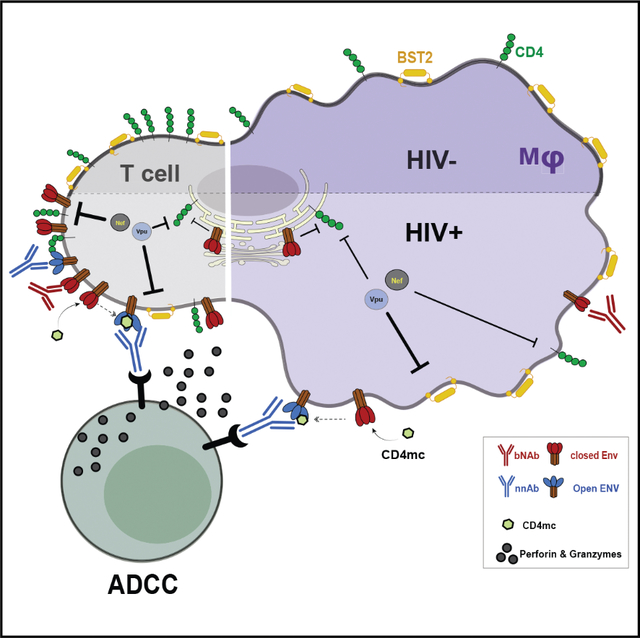
SUMMARY
HIV-1 envelope (Env) conformation determines the susceptibility of infected CD4+ T cells to antibody-dependent cellular cytotoxicity (ADCC). Upon interaction with CD4, Env adopts more “open” conformations, exposing ADCC epitopes. HIV-1 limits Env-CD4 interaction and protects infected cells against ADCC by downregulating CD4 via Nef, Vpu, and Env. Limited data exist, however, of the role of these proteins in downmodulating CD4 on infected macrophages and how this impacts Env conformation. While Nef, Vpu, and Env are all required to efficiently downregulate CD4 on infected CD4+ T cells, we show here that any one of these proteins is sufficient to downmodulate most CD4 from the surface of infected macrophages. Consistent with this finding, Nef and Vpu have a lesser impact on Env conformation and ADCC sensitivity in infected macrophages compared with CD4+ T cells. However, treatment of infected macrophages with small CD4 mimetics exposes vulnerable CD4-induced Env epitopes and sensitizes them to ADCC.
In brief
In this study, Laumaea et al. characterize CD4 downregulation, Env conformation, and ADCC responses in infected macrophages and autologous CD4+ T cells. They report that Nef and Vpu protect infected macrophages from ADCC responses mediated by HIV+ plasma and that small CD4 mimetics sensitize them to ADCC.
Graphical Abstract
INTRODUCTION
Being the only viral protein exposed on the surface of infected cells, the HIV-1 envelope glycoprotein (Env) is the main antigen targeted by neutralizing and non-neutralizing antibodies. Indeed, broadly neutralizing antibodies (bNAbs) have been shown to efficiently mediate antibody-dependent cellular cytotoxicity (ADCC) against productively infected cells.1–4 Most of these studies largely focused on ADCC responses against infected CD4+ T cells. However, comparatively little is known about the conformation of Env at the surface of infected macrophages and consequently its impact on responses against this cell type, with one recent study reporting resistance of infected macrophages to natural killer (NK) cell-mediated killing using select bNAbs.5
Macrophages, which are distributed throughout the body, are highly heterogeneous (reviewed in Qian et al.6). Tissue location of macrophage populations (blood, brain, gut, lungs, liver) is intimately associated with their ontogeny, and several lineages/subsets have been described based on their developmental origins: embryonic yolk sac and fetal liver precursors vs. bone marrow progenitors (reviewed in Ginhoux and Guilliams7). To date the relevance of HIV infection in the different subsets is poorly defined, though evidence support microglia, a tissue-resident subset, as an important target for HIV in the brain.8–10 Despite the susceptibility of macrophages to HIV infection,11–14 there exists important differences between CD4+ T cells and macrophages that could influence the efficiency of eliminating these cells. An important difference is cell surface levels of CD4,15 which have been shown to be important in the exposure of ADCC epitopes on CD4+ T cells.16–18 Interactions of CD4 with functionally conserved residues (365–373) within gp120’s CD4 binding site (CD4BS), in particular, a critical electrostatic interaction between the aspartic acid at position 368 (D368) of gp120 with arginine 59 (R59) of CD4 that modulates infection,19,20 importantly, are key to exposing ADCC epitopes.16,21,22 Once infection is established, CD4 is downregulated from the surface of cells via the accessory proteins Nef and Vpu, as well as Env. These mechanisms are well characterized in CD4+ T cells,16,21,23,24 though there remains a paucity of data on their role in HIV-infected macrophages.
Here we used monocyte-derived macrophages (MDMs) as a model to better understand the role of HIV-1 Nef, Vpu, and Env on CD4 downregulation in this cell type and studied how this impacts Env conformation and susceptibility to ADCC.
RESULTS
HIV-1 mediated CD4 downregulation in macrophages
HIV-1 uses Vpu, Env, and Nef to downregulate CD4 from the surface of primary CD4+ T cells.25–30 To evaluate whether this was also the case for macrophages, which express comparatively little CD4,15 we infected MDMs and autologous CD4+T cells with the wild-type (WT) full-length infectious molecular clone (IMC) of the macrophage tropic, R5 isolate, HIV-1AD8. The relative contribution toward CD4 downregulation by these viral proteins was measured by infecting cells with mutant IMCs unable to express Nef (N–), Vpu (U–) or both accessory proteins (N–U–). Additionally, the impact of Env on CD4 downregulation was measured with IMCs expressing an Env variant containing a mutation in the CD4 binding site (CD4BS) that prevents Env interaction with CD420,21 (D368R; N–U–D368R). In both cell types, the maximal CD4 downregulation was achieved with the WT IMC (Figure 1A; Table S1). Infection with N–, U–, and N–U– but not D368R significantly disrupted CD4 downregulation in primary CD4 T cells. Of note, Env contribution to CD4 downregulation was evident when the D368R mutation was added to the N–U– IMC (N–U–D368R). In agreement with previous observations,16,21 only in this context, infected cells expressed the same levels of CD4 as uninfected cells (Figures 1A and 1B). Neither the abrogation of Nef and/or Vpu nor the CD4BS mutation completely restored CD4 surface levels to those observed in uninfected cells. In both cell types, all three mechanisms of CD4 downregulation (i.e., Nef, Vpu, and Env, N–U–D368R) needed to be disrupted in order to present comparable levels of CD4 expression as mock infected cells (Figures 1A and 1B). As expected, in CD4+ T cells, deletion of Nef or Vpu resulted in a significant increase of cell surface levels of CD4 (Figures 1A and 1B). The same deletions did not impact CD4 levels at the surface of infected macrophages, which remained the same as in WT infected cells. Both Nef and Vpu had to be deleted (N–U–) in order to observe a significant increase in cell surface levels of CD4 in macrophages (Figures 1A and 1B). The lower overall CD4 expression on macrophages, as previously reported15 and supported herein (Figure 1A), likely explains this phenotype. To ensure that this phenotype was not restricted to HIV-1AD8, we performed analogous experiments with two additional R5 (HIV-1JRFL, HIV-1YU2) and one dual tropic (HIV-1CH77) IMCs. Similar phenotypes were observed (Figures 1B and S1E–S1G; Table S1).
Figure 1. CD4 and BST2 downregulation in HIV-1-infected primary CD4+ T cells and autologous macrophages.
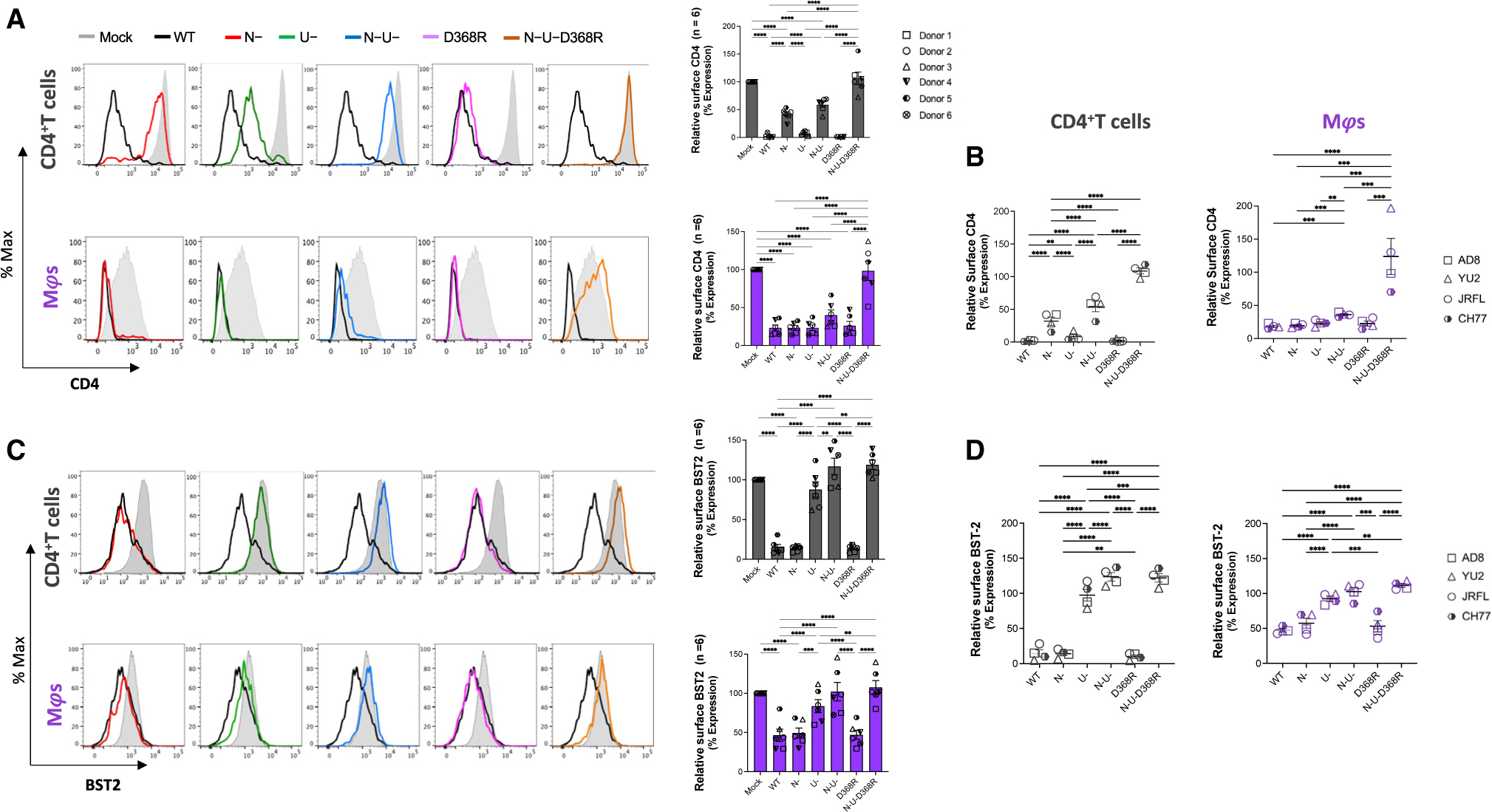
Autologous CD4+ T cells and macrophages were infected with the HIV-1AD8 panel of viruses (WT, Nef-defective [N–], Vpu-defective [U–], or both Nef and Vpu [N–U–] and CD4BS mutant [D368R] and N-U-D368R mutant), and CD4 and BST2 levels were determined 48 h later (CD4+ T cells) or 5 days after infection (macrophages).
(A) Relative surface levels of CD4 on CD4+ T cells and macrophages. Histograms (left) depict representative staining of infected cells and bar graphs (right) show percent fold change in CD4 expression relative to mock (p24+/uninfected cells).
(B) Summary of relative surface levels of CD4 on CD4+ T cells and macrophages infected with a panel of viruses (WT, Nef-defective [N–], Vpu-defective [U–], or both Nef and Vpu [N–U–] and CD4BS mutant [D368R] and N-U-D368R mutant) from HIV-1AD8, HIV-1JR-FL, HIV-1CH77, and HIV-1YU2.
(C) Relative surface expression of BST2 on CD4+ T cells and macrophages. Histograms (left) depict representative staining of infected cells, and bar graphs (right) show percent fold change in BST2 expression of p24+ relative to p24− cells (p24+/p24−).
(D) Summary of relative surface expression of BST2 on CD4+ T cells and macrophages infected with a panel of viruses (WT, Nef-defective [N–], Vpu-defective [U–], or both Nef and Vpu [N–U–] and CD4BS mutant [D368R] and N-U-D368R mutant) from HIV-1AD8, HIV-1JR-FL, HIV-1CH77, and HIV-1YU2. Error bars indicate mean ± SEM. Statistical significance was tested using two-way ANOVA (A and C; n = 6 donors) or mixed effect analysis (B and D; n = 4 viruses) with Holm-Sídák’s multiple comparisons test (*p < 0.05; **p < 0.001; ***p < 0.0001; ns, non-significant).
In HIV-1-infected CD4+ T cells, Nef and Vpu restrict ADCC responses by limiting Env-CD4 complexes that otherwise expose vulnerable epitopes recognized by antibodies commonly present in plasma from infected individuals.16,21,23,31–35 In addition to downregulating CD4, Vpu contributes to protection of infected cells from ADCC by downregulating the restriction factor BST-2, which traps viral particles and results in antigen accumulation at the cell surface.16,21,36,37 We therefore measured cell surface levels of BST-2 in infected CD4+ T cells and autologous macrophages. In both cell types, Vpu was the principal viral determinant modulating BST-2 expression (Figure 1C). Similar results were obtained with the other IMCs tested (Figures 1D and S1E–S1G and Table S1). Thus, with respect to BST-2 modulation, Vpu performs parallel roles on CD4+ T cells and macrophages. Of note, we observed that for HIV-1 AD8 and YU2, full recovery of BST-2 levels at the cell surface required Nef deletion (N–U–), in agreement with a recent report suggesting that Nef from some HIV-1 strains, including AD8, can downregulate BST-2.38
Env conformation at the surface of infected primary CD4+ T cells and autologous macrophages
Using a panel of anti-Env antibodies (Table S2) and IMC variants (WT, N-, U-, N-U-, D368R, and N-U-D368R), we evaluated Env conformation at the surface of infected macrophages and autologous CD4+ T cells. In both cell types, recognition of infected cells with bNAbs having a conformational preference for the “closed” trimer (PG9, PGT121, 10.1074, 3BNC.117, PGT151) was modulated by Vpu. Vpu deletion (U–) alone or in combination with Nef (N–U–) enhanced recognition, in a manner that reflected overall amount of Env at the surface of infected cells, as measured with the conformational independent 2G12 antibody (Figure 2A). This is consistent with previous studies demonstrating that Vpu-mediated BST-2 downregulation prevents virion accumulation on the cell surface, thereby reducing the overall levels of detectable Env.16,21,36,37,39 The impact of Vpu on cell recognition by the different bNAbs was markedly lower on infected macrophages compared with CD4+ T cells (Figure 2).
Figure 2. Env recognition at the surface of HIV-1-infected primary CD4+ T cells and autologous macrophages by a panel of bNAbs.
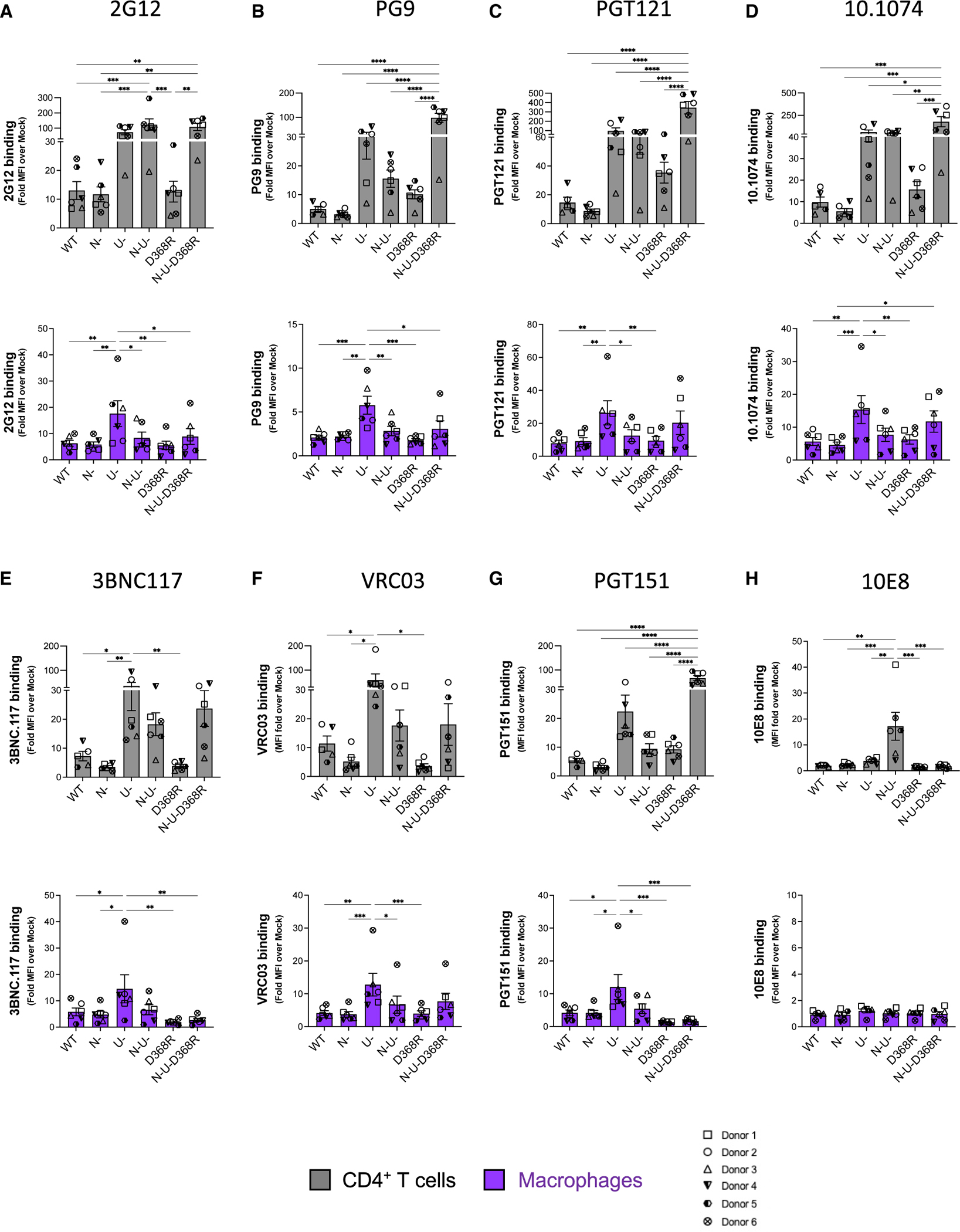
Staining of CD4+ T cells and macrophages infected with HIV-1AD8 panel of viruses (WT, Nef-defective [N–], Vpu-defective [U–], or both Nef and Vpu [N–U–] and CD4BS mutant [D368R] and N-U-D368R mutant) with (A) 2G12 a conformation independent antibody; (B) the V1V2-apex antibody PG9; V3glycan antibodies (C) PGT121 and (D) 10.1074; CD4BS antibodies (E) 3BNC117 and (F) VRC03; (G) PGT151, which targets the gp120-gp41 interface; and the (H) anti-MPER antibody 10E8. Values represent fold change in MFI relative to mock. Error bars indicate mean ± SEM.Statistical significance was tested using two-way ANOVA with Holm-Šídák’s multiple comparisons test (*p < 0.05; **p < 0.001; ***p < 0.0001; ns, non-significant). Shown are data from 6 donors.
PG9 recognizes V1V2 epitopes on the trimer apex40–42 and preferentially interacts with the “closed” Env. Indeed, interaction of Env with membrane-bound CD4 was shown to decrease PG9 interaction.16 Supporting the existence of a “closed” conformation of the unliganded trimer, we observed similar levels of Env recognition by PG9 (Figure 2B) at the surface of WT infected CD4+ T cells and macrophages (Figure 2B). Interestingly, while CD4-Env disruption in the context of the N-U-D368R infected cells significantly enhanced binding of PG9 on CD4+ T cells, this was not observed for macrophages (Figure 2B, lower panel). This phenotype was recapitulated with the HIV-1CH77 and HIV-1YU2 N-U-D368R mutants (Figure S2B).
Recent work showed comparable staining of Env with the 3BNC117 CD4BS Ab on both cell types,5 while others such as N65 and NIH-45–4643 showed better recognition of macrophages in comparison to CD4+ T cells. Consistent with Clayton et al.,5 we observed that 3BNC117 and VRC03 recognize Env similarly on both WT infected CD4+T cells and macrophages (Figures 2E and 2F). These CD4BS antibodies displayed improved recognition of cells infected with a Vpu- virus (Figures 2E and 2F). This recognition was diminished by deleting Nef. In the absence of Nef, there is more CD4 on the cell surface interacting with Env,16 therefore occluding the CD4BS. The D368R variant slightly decreased recognition by these antibodies, as expected due to the role played by D368 for their interaction.44,45 While disruption of the Env-CD4 interaction in the absence of Nef and Vpu (N–U– D368R) enhanced 3BNC117 recognition of infected CD4+ T cells, this did not happen in macrophages. This is likely due to low amounts of CD4 on the surface of macrophages, which might not be sufficient to compete with this CD4BS bNAb (Figures 2E and 2F). When PGT151, an antibody that recognizes the interface between gp120 and gp41,46 was tested, we observed efficient recognition of cells infected with a virus lacking Vpu or expressing Env D368R, but not when both Vpu and Nef were absent (Figure 2G). At the surface of infected cells, CD4 and Env are interacting on the same membrane, and the CD4 domains D3–D4 may prevent PGT151 access of its epitope on Env, which is located beneath the CD4 binding site. As observed for 3BNC117, disruption of Env-CD4 interaction in absence of Nef and Vpu (N–U– D368R) enhanced the recognition of infected CD4+ T cells by PGT151 but not in infected macrophages, consistent with low levels of CD4 in this cell type.
The bNAb 10E8 recognizes the membrane proximal external region (MPER) of gp41.47 While binding the unliganded trimer likely explains its neutralization breath,47 10E8’s epitope, as with other MPER Abs, is better exposed upon CD4 interaction.48 Accordingly, Nef deletion enhanced recognition of CD4+ T cells infected with the N– and N–U– constructs and was dependent on Env-CD4 interaction since introduction of the D368R mutation significantly decreased its binding (Figure 2H). Interestingly, no 10E8 recognition for macrophages was observed upon Nef and Vpu deletion (Figure 2H) despite enhanced recognition of CD4+T cells infected with the N- and N-U- constructs. For CD4+ T cells, this effect appeared to be dependent on Env-CD4 interaction since introduction of the D368R mutation significantly decreases its binding (Figure 2H). Whether this cell-type-dependent discrepancy is linked to the different levels of CD4 at the cell surface remains to be determined. Of note, however, similar phenotypes were observed with the three additional IMCs tested (JRFL, YU2, and CH77) (Figures S2–S4).
We then evaluated the recognition of infected cells by nnAbs. In agreement with the “closed” conformation adopted by the unliganded Env trimer, none of the tested nnAbs efficiently recognized WT infected cells (Figures 3A–3D). The CD4-triggerable nature of the epitope they recognize has been well documented.16–18,23,31,49,50 Accordingly, all Abs recognized more efficiently N–, U–, and N–U– infected CD4+ T cells, with the latter two mutants (U–, N–U–) to a greater magnitude, owing also to unconstrained action of BST-2 (Figure 3A). Abrogation of Env-CD4 interaction by introduction of the D368R mutation significantly impaired recognition by these nnAbs (Figure 3A), further supporting their CD4-induced nature. Importantly, consistent with lower CD4 levels on macrophages,15 these Abs barely detected infected macrophages with the exception of N– and N–U– infected cells, albeit to a lesser magnitude than CD4+ T cells Figure 3B).
Figure 3. Recognition of HIV-1-infected primary CD4+ T cells and autologous macrophages by non-neutralizing antibodies.
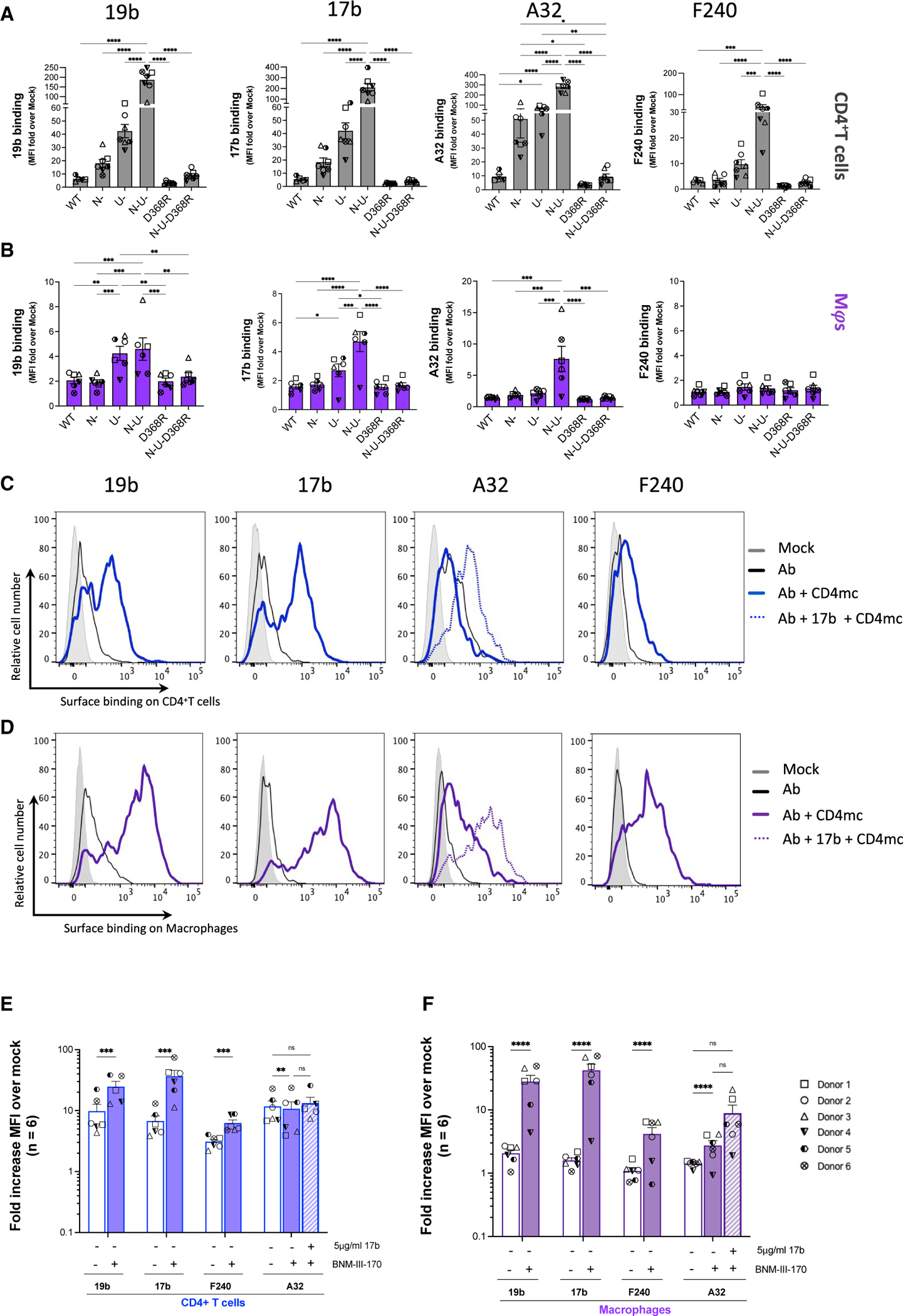
Autologous (A and C) CD4+ T cells and (B and D) macrophages infected with the HIV-1AD8 panel of viruses (WT, Nef-defective [N–], Vpu-defective [U–], or both Nef and Vpu [N–U–] and CD4BS mutant [D368R] and N–U–D368R mutant) for 48h (CD4+ T cells) or 5 days (macrophages) were stained with non-neutralizing antibodies 19b (V3 crown), 17b (co-receptor binding site), A32 (anti-cluster A), and F240 (gp41-disulfide loop region). Histograms depict representative staining of infected cells of 19b, 17b, F240, and A32 with or without CD4mc BNM-III-170 or A32 with 17b and CD4mc. Fold increase in nnAb binding for CD4+ T cells (E) and macrophages (F), relative to mock/uninfected cells, in the presence of the CD4mc is shown on the y axis. Error bars indicate mean ± SEM. Statistical significance was tested using two-way ANOVA with Holm-Šídák’s multiple comparisons test (A and B) or mixed effect analysis (E and F). (*p < 0.05; **p < 0.001; ***p < 0.0001; ns, non-significant). Shown are data from 6 donors.
Exposing CD4i epitopes on the surface of infected macrophages
It has been well documented that nnAbs fail to recognize Env in its “closed” conformation.1,3,16,21–24,39,51,52 As shown in Figure 3, infected macrophages make no exception with poor recognition of WT infected cells by all tested nnAbs.
Small CD4-mimetic compounds (CD4mc) were shown to expose vulnerable Env epitopes at the surface of infected CD4+ T cells, resulting in their sensitization to ADCC mediated by nnAbs and plasma from infected individuals.22,52,53 To evaluate whether this could apply to infected macrophages, we used BNM-III-170, a CD4mc extensively used to expose epitopes recognized by nnAbs on infected primary CD4+ T cells.53–56 To facilitate comparisons, we infected autologous primary CD4+ T cells and evaluated the capacity of four non-neutralizing antibodies to recognize infected cells (macrophages and CD4+ T cells). As described above, in the absence of CD4mc the four nnAbs tested (19b, 17b, A32, and F240) poorly recognized WT infected macrophages or CD4+ T cells (Figures 3A and 3B). Consistent with the capacity of BNM-III-170 to “open up” Env and expose vulnerable epitopes, CD4mc addition enabled efficient recognition by 19b, 17b, and F240 (Figures 3C and 3D). In both infected CD4+ T cells and macrophages, and in agreement with the literature,52,53,55,56 efficient exposure of the gp120 inner domain cluster A region recognized by A32 required addition of the CoRBS 17b antibody together with the CD4mc, being more notable with infected MDMs (Figures 3C–3F and S4), potentially highlighting the efficiency of the CD4mc on MDMs that is otherwise masked by the CD4-rich surface of CD4+ T cells so that only a slight increase in A32 binding from baseline is observed when CD4mc and/or 17b are added. This phenotype was recapitulated with the other IMCs (Figures S4D and S4E).
BNM-III-170 sensitizes HIV-infected macrophages to ADCC mediated by HIV+ plasma
A recent study reported that infected macrophages are resistant to ADCC mediated by NK cells.5 However, it remained to be determined whether CD4mc renders infected macrophages susceptible to ADCC. To do this, we adapted our fluorescence-activated cell sorting (FACS)-based ADCC assay that uses infected CD4+ T cells3,16 for assessment of killing of infected macrophages (Figure 4, gating strategy shown in Figure S5). Briefly, macrophages differentiated for 7 days were infected with HIV-1AD8 WT and virus defective for Nef and Vpu (N–U–) for 5 days. WT infected cells were then treated with or without BNM-III-170. N–U– infected cells were used as a positive control for ADCC activity mediated by nnAbs. ADCC activity was then measured following 5 h incubation with autologous peripheral blood mononuclear cells (PBMCs) as effector cells in the presence of plasma from nine different HIV-1 infected individuals. Consistent with their role in protecting infected CD4+ T cells from ADCC, deletion of Nef and Vpu (N–U–) markedly increased their susceptibility to ADCC (Figure 4A). However, in infected macrophages, deletion of Nef and Vpu only led to a small increase in ADCC susceptibility (Figure 4B). This is consistent with the modest increase in CD4 levels at the surface of N-U-compared with WT infected macrophages (Figure 1A). Of note, addition of BNM-III-170 sensitized both infected CD4+ T cells and macrophages to ADCC mediated by plasma from nine different HIV-1-infected individuals (Figure 4). To determine the contribution of the different effectors (Figure S6) within the PBMC pool in mediating ADCC, we adapted the above described ADCC assay using NK cells or monocytes as effectors, with plasma from five different donors. In addition, the bNAbs that stained both CD4+ T cells and macrophages similarly (3BNC117, PGT121, 10.1074, and 2G12) and had been shown to mediate ADCC of CD4+ T cells (3BNC117 and PGT121)5 were selected to assess ADCC functionality for both CD4+ T cells and macrophages in the context of NK cells and monocytes.
Figure 4. Small CD4mc BNM-III-170 sensitizes HIV-1-infected macrophages to ADCC mediated by HIV+ plasma.
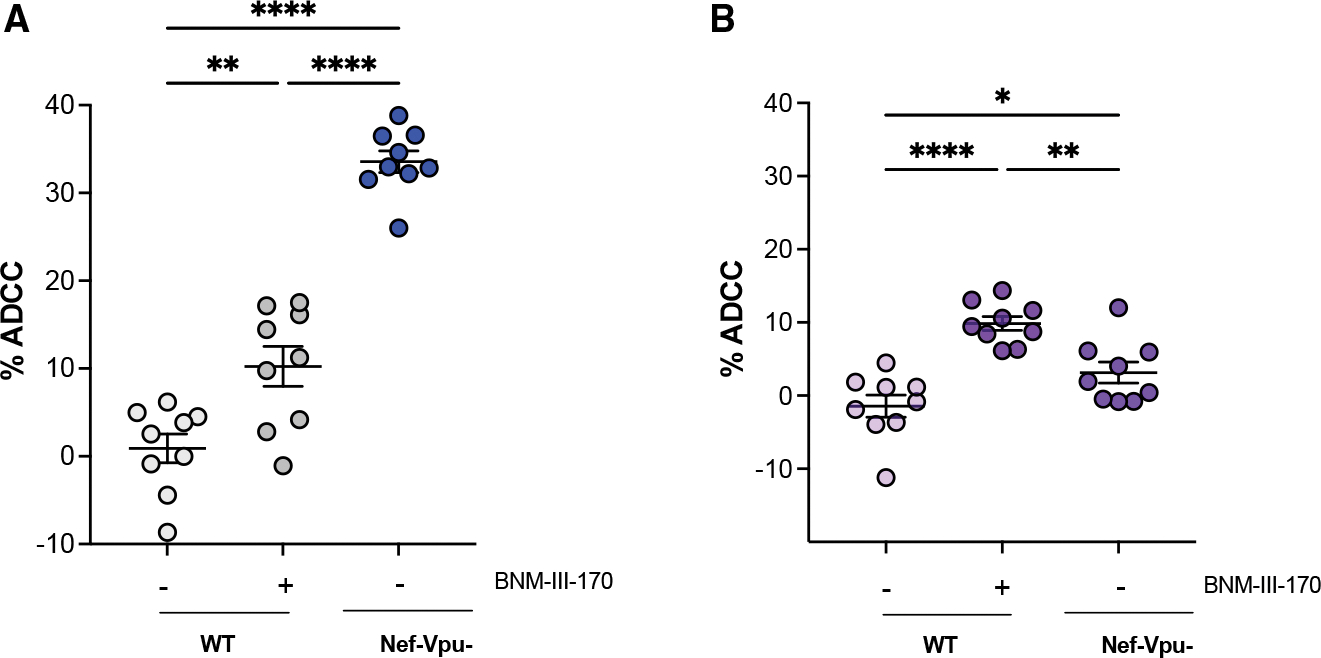
Autologous CD4+ T cells and macrophages were infected with HIV-1AD8 (WT and Nef- and Vpu-defective [N–U–] virus) and subsequently incubated with autologous PBMCs for 5 h in the presence of HIV+ plasma with or without the CD4mc BNM-III-170.
Percent (%) ADCC of CD4+ T cells (A) and macrophages (B). Error bars indicate mean ± SEM. Statistical significance was tested using ordinary one-Way ANOVA followed by Holm-Šídáks multiple comparisons test (*p < 0.05; **p < 0.001; ***p < 0.0001; ns, non-significant). Shown are data from at least three donors.
Results suggest that CD4+ T cells are susceptible to killing by NK cells (Figure S7C) in the presence of the CD4mc consistent when PBMCs are effectors (Figures 4 and S7B) and that absence of Nef and Vpu further enhances susceptibility to NK-mediated killing. Absence of Nef and Vpu also increased susceptibility of infected CD4+ T cells to killing by monocyte effectors (Figure S7D), but addition of CD4mc did not facilitate killing of WT infected CD4+ T cells by monocytes. In contrast, while PBMCs maintained killing of macrophages in the presence of the CD4mc (Figure S7E), NK cells were inefficient even with the addition of the CD4mc (Figure S7F). Interestingly, monocytes exhibited potent ADCC of WT infected macrophages in the presence of the CD4mc (Figure S7G). Furthermore, macrophages infected with the mutant N–U– were not susceptible to monocyte-mediated killing, displaying a phenotype similar to Figure 4B.
Further analysis to assess the ability of bNAbs to mediate ADCC of infected macrophages suggests that, despite recognition of Env by the bNAbs (Figure 2) utilized here, these antibodies appeared to be less efficient, relative to HIV+ plasma in the presence of the CD4mc, at mediating ADCC of macrophages by the different effectors (Figures S7H–S7J), compared to when CD4+ T cells were used as targets. However, whether this relates to a differential capacity of HIV+ plasma (in the presence of CD4mc) and bNAbs to crosslink FcγR on effector cells remains to be determined. Finally, these results also show a differential susceptibility of infected CD4+ T cells and macrophages to NK cells and monocyte-mediated ADCC.
Integrated analysis of the associations between recognition of Env epitopes CD4 and BST-2 on the surface of macrophages and CD4+T cells
To assess the association of Env epitope recognition, CD4 and BST-2, we performed a network correlation analysis, done separately for CD4+ T cells and macrophages (Figure 5). For macrophages, the network had two distinct clusters of significant correlations (Figure 5B), whereas the T cell network was more intertwined (Figure 5A). While there were significant associations between nnAb recognition in the presence of the CD4mc and the measurements of BST-2 and CD4, only two antibodies, A32 and F240, significantly correlated with BST-2 measurements (Figure 5B, bottom panel). Importantly there was a striking disconnect of associations between bNAb binding and measurements of CD4, suggesting that Env epitope exposure on macrophages is minimally influenced by the surface levels of CD4. In stark contrast, the correlation network for CD4+ T cells (Figure 5A, top panel) was more balanced with intricate relationships between the levels of CD4/BST-2 and Env recognition by all antibodies, including the nnAbs. Furthermore, the inverse correlations between the levels of CD4 and recognition by nnAbs in the presence of the CD4mc on CD4+ T cells was more pronounced (Figure 5A, bottom panel), supporting that the levels of CD4 on CD4+ T cells greatly influence epitope recognition. Importantly, the heatmap analysis (Figure 5C) highlights that the differences between macrophages and CD4+ T cells are not virus strain dependent.
Figure 5. Network associations of Env epitope recognition with BST2 and CD4 on CD4+ T cells and macrophages.
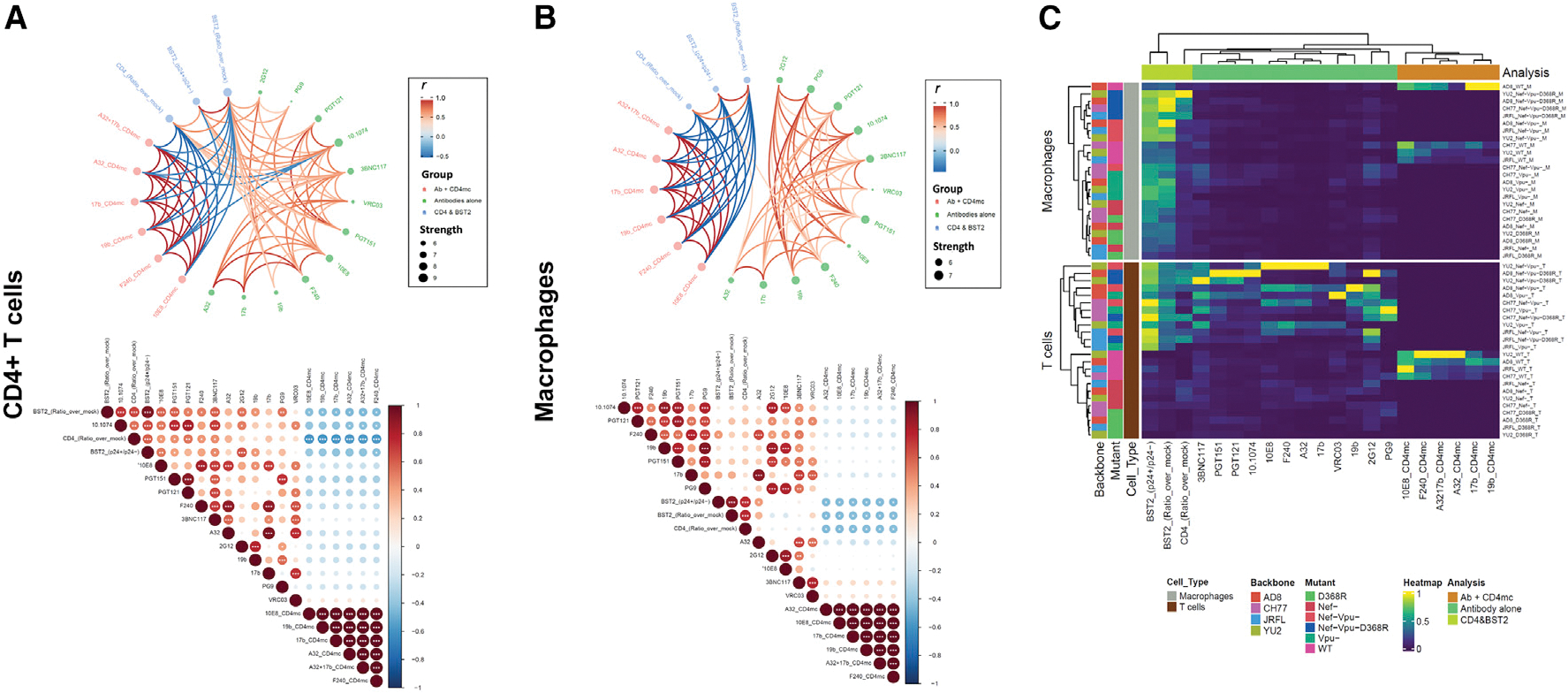
(A and B) Circular edge bundling plots (top) and correlation matrices (bottom) for CD4+ T cells (A) and macrophages (B) where red and blue edges (top) and circles (bottom) represent positive and negative pairwise correlations between connected variables, respectively. Nodes in the edge bundling plots (top) are color-coded based on the grouping of variables shown in the legend with node size corresponding to degree of relatedness of correlations. Only significant correlations (p < 0.05, Spearman rank test) are displayed in the edge bundling plots (top). In the correlograms (bottom), *p < 0.05, **p < 0.01, ***p < 0.005.
(C) Heatmap showing Env, BST, and CD4 expression relative to virus strains tested and cell type. Columns represent recognition of BST2, CD4, and Env clustered based on similarity and grouped by analysis method according to the color code provided. Rows represent the viruses/mutants grouped according to cell type. Virus IDs are clustered according to their binding profile and done separately for each cell type.
DISCUSSION
Recent work reported differential recognition of Env epitopes on the surface of infected macrophages5,43 with one study highlighting strong recognition of CD4BS epitopes on the surface of infected macrophages compared with CD4+ T cells.5 Here, we report that similarly to infected primary CD4+ T cells, Env on macrophages is predominantly in a “closed” conformation.
HIV-1 uses three different proteins (Nef, Vpu, and Env) to efficiently downregulate CD4 from the cell surface. While Nef targets CD4 molecules already present at the plasma membrane, Vpu and Env target newly synthesized CD4 for degradation. While in both cell types deletion of Nef and Vpu and abrogation of Env-CD4 interaction (N-U-D368R) was required to restore CD4 levels to those observed with uninfected cells, the effect of Nef and Vpu appeared more prominent in CD4+ T cells. Indeed, in this cell type but not in macrophages, their individual deletion (N– or U–) significantly increased CD4 levels. The relatively limited amount of CD4 at the surface of macrophages compared with autologous CD4+ T cells (Figure 1A and Lee et al.15) might facilitate the task of the two other remaining viral proteins to fully downregulate CD4 in macrophages.
In agreement with previous studies,16–18 we found that the ability of Nef and Vpu to protect infected cells from ADCC mediated by HIV+ plasma was linked to CD4 expression. Both CD4+ T cells and macrophages infected with the WT virus were resistant to ADCC. However, while Nef and Vpu deletion dramatically increased the susceptibility of infected CD4+ T cells to ADCC, it only had a minor effect in infected macrophages (Figures 4 and S7). This is consistent with the limited amount of CD4 present at the surface of infected macrophages to “open-up” Env and expose ADCC vulnerable epitopes. Nevertheless, similarly to infected primary CD4+ T cells, we were able to expose vulnerable epitopes at the surface of infected macrophages using the CD4mc BNM-III-170. Of note, exposure of the gp120 inner domain A32 epitope further required the combination of the CD4mc with a CoRBS Ab such as 17b. This combination was recently shown to stabilize an ADCC vulnerable conformation of Env, State 2A,53 indicating that this conformation can also be stabilized on infected macrophages. Interestingly, the combination of nnAbs (17b + A32) and CD4mc was also recently found to decrease HIV-1 replication and reduce HIV DNA in CD4+ T cells in humanized mice.56 Accordingly, the data herein demonstrate that the addition of BNM-III-170 improved recognition and ADCC susceptibility of HIV-1 infected macrophages by nnAbs, commonly elicited antibodies that predominate in HIV+ plasma. These findings warrant further efforts to test whether CD4mc could enable nnAbs to eliminate infected macrophages in vivo.
Since a recent study reported that macrophages are resistant to NK cell killing,5 we decided to perform our ADCC assay using PBMCs (Figure 4), NK cells (Figure S7), and monocytes (Figure S7) as effector cells. Our results suggest that infected macrophages could be sensitive to Fc-effector functions mediated by monocytes but not NK cells. Consistent with published literature,5 we observed that NK cells are inefficient at eliminating HIV-infected macrophages even when bNAbs or potent ADCC plasma are used. Interestingly monocytes demonstrate efficient killing capacity in the presence of the CD4mc (Figure S7). Monocytes as efficient mediators of ADCC have previously been explored. Earlier work suggested that CD16+ monocytes were able to efficiently perform ADCC of tumor cells,57 HSV-infected cells,58 SARS-CoV-2-infected cells,59 and relevant to this study, HIV-1-infected CD4+ T cells.60 It is possible that monocytes may bear the appropriate markers required for efficient targeting and elimination of HIV-infected macrophages compared with NK cells. Further work to scrutinize the mechanisms involvedis therefore warranted.
Here, we used MDM as a model, and it would therefore be important to ascertain whether the phenotypes observed herein are consistent with other cells of the myeloid lineage, in particular microglia, an important HIV reservoir within the central nervous system (CNS).8 While we show that the small molecule CD4mc BNM-III-170 was able to expose CD4i epitopes on infected macrophages permitting ADCC, we acknowledge that the CNS may still remain inaccessible.
Altogether, our data provide further insights on the role of viral proteins in modulating cell-surface CD4 and Env conformation on infected macrophages, as well as proof of principle that HIV-1-infected macrophages can be sensitized to recognition and ADCC by CD4i antibodies. These findings provide additional information for the development of immunotherapies aimed at targeting and eliminating the HIV-1 reservoir in vivo.
Limitations of the study
Given that our study suggests differences in Env conformation at the surface of infected macrophages and CD4+ T cells, it is therefore possible that similar differences might exist on Env incorporated in viral particles derived from these cell types. Assays such as the one developed by Ding and colleagues61 might be useful in that regard. Additionally, here we used MDM as a model, and it would be important to extend our findings to additional macrophage subtypes from different lineages. Nevertheless, the data presented here using MDMs facilitate important assertions as to the mechanism of macrophage evasion of immune responses.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrés Finzi (andres.finzi@umontreal.ca).
Materials availability
All reagents generated in this study are available from Andrés Finzi (andres.finzi@umontreal.ca) with a completed Materials Transfer Agreement.
Data and code availability
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics statement
Written informed consent was obtained from all study participants and research adhered to the ethical guidelines of CRCHUM and was reviewed and approved by the CRCHUM Institutional Review Board (ethics committee, approval number CE16.164-CA). Research adhered to the standards indicated by the Declaration of Helsinki. All participants were adult and provided informed written consent prior to enrollment in accordance with Institutional Review Board approval.
Experimental models: Cell lines
The HEK293T human embryonic kidney cells (293T) (obtained from ATCC and the NIH AIDS Research and Reference Reagent Program, respectively) were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 5% FBS (VWR) and 100 mg/ml of penicillin-streptomycin (Wisent).
Experimental models: Primary cells
Primary cells were grown as previously described.16 Briefly cryopreserved human peripheral blood mononuclear cells (PBMCs) isolated by ficoll density gradient from 6 healthy donors (HIV and hepatitis C virus [HCV] seronegative); 4 males (32, 53, 64 and 67 years of age) and 2 females (45 and 46 years of age), who gave written informed consent under research protocols approved by the CRCHUM, were thawed and monocytes were isolated by plate adherence in 10cm petri dishes (Sarsdedt) for 1h in Iscove’s modified Dulbecco medium (IMDM). Non-adherent cells were collected while adherent cells washed extensively in serum free media and allowed to differentiate to macrophages for seven days in IMDM supplemented with 100 mg/ml of penicillin-streptomycin and 10% pooled human sera (Valley Biomedicals), with a half media change at day 3. CD4+ T cells were isolated from the non-adherent cells by negative selection (EasySep) activated and maintained in culture as previously described.52,74 NK cells and monocytes were isolated from PBMCs using the EasySep Human NK Cell isolation Kit (Stemcell Technologies, Cat #17955) and EasySep Human Monocyte enrichment kit (Stemcell Technolocies, Cat $19059) respectively. PBMC, NK and monocytes effectors were subsequently rested overnight at 37°C in culture medium prior to ADCC assays.
METHOD DETAILS
Bacterial and virus strains
The infectious molecular clones (IMCs) of HIV-1 strain AD8: (pAD8+) HIV-1AD866 Vpu- (AD8-U-), Nef- (AD8-N-) and Nef-Vpu- (AD8-N-U-); and HIV-1 YU2 strain: HIV-1 YU2,67 Vpu- (YU2-U-) and Nef-Vpu- (AD8-N-U-). The D368R derivatives of AD8 and YU2 and Nef defective YU2 (YU2-N-) were generated by mutagenesis using primers listed in key resources table. The IMCs of HIV-1 strains JRFL was kindly provided by Dr Dennis Burton. The Nef-defective and Vpu-defective JR-FL IMCs were previously described.17 The CH77 (CH077) transmitter founder was previously described.68 Mutated IMCs were previously described.22,68,75–77 Site-directed mutagenesis was performed on JR-FL and CH77 IMCs to introduce the Env D368R mutation using the QuikChange II XL site-directed mutagenesis protocol. The presence of the desired mutations was determined by automated DNA sequencing.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-gp12- outer domain 2G12 | NIH Reagents program | N/A |
| anti-CD4 binding site VRC03 | NIH Reagents program | Cat # 12032 |
| anti-CD4 binding site 3BNC117 | Scheid et al.62 | Cat #: 12474 |
| anti-V2 apex PG9 | Polymum Scientific, Klosterneuburg, Austria | N/A |
| anti-V3 10.1074 | Dr. Michel Nussenzweig; Mouquet et al.63 | N/A |
| anti-V3 PGT121 | IAVI; Walker et al.64 | Cat # 12343 |
| anti-gp120-gp41 interface PGT151 | IAVI; Falkowska et al.46 | N/A |
| anti-MPER 10e8 | NIH Reagents program; Huang et al.47 | Cat# 12294 |
| anti-gp41 DSR F240 | NIH Reagents program; Cavacini et al.65 | Cat# 7623 |
| anti-V3 crown 19b | NIH Reagents program | N/A |
| anti-coreceptor binding site 17b | NIH Reagents program | Cat # 4091 |
| anti-cluster A A32 | NIH Reagents program | Cat # 11438 |
| PE-conjugated anti-HIV p24 (Clone KC57-RD1) | Beckman Coulter | Cat# CO6604667 |
| FITC-conjugated anti-HIV-p24 (Clone KC57) | Beckman Coulter | Cat# 6604665 |
| BUV395 conjugated anti-CD3 (Clone UCHT1) | BD Biosciences | Cat# 563546 |
| BV650 conjugated anti-CD11b (Clone: M1/70) | Biolegend | Cat# 101239 |
| PE-Cy7 conjugated anti-BST2(Clone RS38E) | Biolegend | Cat# 348416 |
| BV421 conjugated anti-CD4 (Clone OKT4) | Biolegend | Cat# 317434 |
| BV650 conjugated anti-CD19 (CloneHIB19) | Biolegend | Cat# 302201 |
| FITC conjugated anti-CD16 (Clone 3G8) | BD Pharmingen | Cat # 555406 |
| PerCPCy5.5conjugate anti-CD14 (Clone M5E2) | BD Pharmingen | Cat # 550787 |
| PE conjugated anti-CD56 (Clone NCAM-1) | BD Pharmingen | Cat # 555516 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| HIV-1 AD8 (Proviral DNA) | Theodore et al.66 | Genbank Accession no. AF004394.1 |
| HIV-1 YU2 (Proviral DNA) | Li et al.67 | N/A |
| HIV-1 JRFL (Proviral DNA) | Dr. Dennis Burton | N/A |
| HIV-1 CH77 T/F (Proviral DNA) | Ochsenbauer et al.68 | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Plasma from HIV-1-infected individuals | FRQS AIDS network | N/A |
| Human PBMC from uninfected individuals | FRQS AIDS network | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| CD4mimetic BNM-III-170 | Dr. Amos B. Smith III; Melillo et al.69 | N/A |
| Dimethyl sulfoxide (DMSO) | Thermo Fisher Scientific | Cat# BP2311 |
| Phytohemagglutinin-L (PHA-L) | Sigma | Cat# L2769 |
| Dulbecco’s modified Eagle’s medium (DMEM) | Wisent | Cat# 319-005-CL |
| RPMI 1640 medium | Thermo Fisher Scientific | Cat#11875093 |
| Penicillin/streptomycin | Wisent | Cat# 450-201-EL |
| Fetal bovine serum (FBS) VWR Cat# 97068-085 | VWR | Cat# 97068-085 |
| Phosphate buffered saline (PBS) | Wisent | Cat# 311-010-CL |
| Tris-buffered saline (TBS) | Thermo Fisher Scientific | Cat# BP24711 |
| Bovine Serum Albumin (BSA) | BioShop | Cat# ALB001.100 |
| Formaldehyde-37% | Thermo Fisher Scientific | Cat# F79-500 |
| Cell proliferation dye eFluor670 | eBioscience | Cat# 65084085 |
| Cell proliferation dye eFluor450 | eBioscience | Cat# 65084285 |
| LIVE/DEAD Fixable AquaVivid Cell Stain | Thermo Fisher Scientific | Cat# L43957 |
| Tween 20 | Thermo Fisher Scientific | Cat# BP337-500 |
| Fc Blocking Reagent (human) | Miltenyi | Cat# 130-059-901 |
| Pooled human sera | Valley Biomedicals | HS1004C |
| Iscove’s modified Dulbecco medium (IMDM) | Gibco-ThermoScientific | Cat# 12440053 |
|
| ||
| Critical commercial assays | ||
|
| ||
| EasySep human CD4+ T cell enrichment kit | Stem Cell Technologies | Cat# 19052 |
| EasySep human NK cell isolation kit | StemCell Technologies | Cat# 17955 |
| EasySep human Monocyte enrichment kit | StemCell Technologies | Cat# 19059 |
| Cytofix/Cytoperm Fixation/Permeabilization Kit | BD Biosciences | Cat# 554714 |
| Mix-n-Stain CF-647 Antibody Labeling Kit (50–100μg) | Sigma-Aldrich | MX647S100-1KT |
| BD PhosFlow Perm/Wash Buffer I | BD Biosciences | Cat# 557885 |
| QuikChange II XL Site directed mutagenesis Kit | Agilent Technologies | Cat # 200522 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T human embryonic kidney cells | ATCC | CAT # CRL-3216 |
|
| ||
| Oligonucleotides | ||
|
| ||
| ENV-YU2-D368R-FWD:5′CCTCAGGAGGGCGACCAGAAATT GTAAC | Integrated DNA Technologies (IDT) | N/A |
| ENV-YU2-D368R-REV:5′GTTACAATTTCTGGTCGCCCTCC TGAGG | Integrated DNA Technologies (IDT) | N/A |
| ADAD368RFWD:CAATCCTCAGGAGGGCGCCCAGAAATTGTAATG | Integrated DNA Technologies (IDT) | N/A |
| ADAD368RREV:CATTACAATTTCTGGGCGCCCTCCTGAGGATTG | Integrated DNA Technologies (IDT) | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pAD8+ (IMC of HIV-1 AD8 WT) | Theodore et al.66 | GenBank accession no. AF004394.1 |
| HIV-1 AD8-Nef- (Proviral DNA) | Dr. Frank Kirchoff | N/A |
| HIV-1 AD8-Vpu- (Proviral DNA) | Krapp et al.70 | N/A |
| HIV-1 AD8-Nef-Vpu- (Proviral DNA) | Dr. Frank Kirchoff | N/A |
| HIV-1 AD8-D368R (Proviral DNA) | This paper | N/A |
| HIV-AD8-Nef-Vpu-D368R (Proviral DNA) | This paper | N/A |
| HIV-1 YU2-Nef- (Proviral DNA) | This paper | N/A |
| HIV-1 YU2-Vpu- (Proviral DNA) | Dr. Frank Kirchoff | N/A |
| HIV-1 YU2-Nef-Vpu- (Proviral DNA) | Dr. Frank Kirchoff | N/A |
| HIV-1 YU2-D368R (Proviral DNA) | This paper | N/A |
| HIV-YU2-Nef-Vpu-D368R (Proviral DNA) | This paper | N/A |
| HIV-1 JRFL-Nef- (Proviral DNA) | Prévost et al.17 | N/A |
| HIV-1 JRFL-Vpu- (Proviral DNA) | Prévost et al.17 | N/A |
| HIV-1 JRFL-Nef-Vpu- (Proviral DNA) | Prévost et al.17 | N/A |
| HIV-1 JRFL-D368R (Proviral DNA) | Ding et al.71 | N/A |
| HIV-JRFL-Nef-Vpu-D368R (Proviral DNA) | This paper | N/A |
| HIV-1 CH77 -Nef- (Proviral DNA) | Prévost et al.17 | N/A |
| HIV-1 CH77 -Vpu- (Proviral DNA) | Kmiec et al.72 | N/A |
| HIV-1 CH77 -Nef-Vpu- (Proviral DNA) | Ding et la.23 | N/A |
| HIV-1 CH77 -D368R (Proviral DNA) | This paper | N/A |
| HIV-CH77 -Nef-Vpu-D368R (Proviral DNA) | This paper | N/A |
| Vesicular Stomatitis virus G (VSV-G) plasmid | Emi et al.73 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo software v10.4 FlowJO, | LLC http://docs.flowjo.com/vx/ | N/A |
| Prism software Graphpad v9.3.0 | https://www.graphpad.com/scientificsoftware/prism/ | N/A |
| Adobe Illustrator Version 26.31 | https://www.adobe.com/products/illustrator | N/A |
|
| ||
| Other | ||
|
| ||
| BD FORTESSA Flow Cytometer | BD Biosciences | N/A |
| Clear V-bottom 96 well plates (cell culture treated) | Corning | Cat #0877126 |
Antibodies and HIV + plasma
The following antibodies were used to assess cell surface staining: 2G12, PG9, PGT121, 10.1074, 3BNC117, VRC03, PGT151, 10e8, 19b, 17b, A32, F240. Details of which are listed in the key resources table. The panel of anti-HIV antibodies were conjugated with CF647 probe (Sigma Aldrich) as per the manufacturer instructions and used for cell-surface staining of HIV-1-infected primary CD4+ T cells and macrophages. Mouse anti-human CD4 (Clone OKT4, BV421-conjugated; Biolegend, San Diego, CA, USA) and mouse anti-human BST2 (clone RS38E, PE-Cy7-conjugated; BioLegend, San Diego, CA, USA) were used as primary antibodies for cell surface staining. To confirm purity of macrophages mouse anti-human CD3 (Clone UCHT1; BUV395 conjugated; BD Biosciences), and to confirm differentiation of monocyte differentiation to macrophages the Rat anti-human/mouse CD11b (BV650 conjugated; Biolegend) were also used. To confirm purity of enriched monocytes and NK cells and to determine cell subsets in PBMC, monocyte and NK effector fractions, cells were stained with mouse anti-human CD3 and mouse anti-human CD19 (Clone HIB19; BV650 conjugated; Biolegend) to exclude lymphocytes. While the mouse anti-human CD14 (Clone M5E2; PerCPCy5.5 conjugated; BD Pharmingen) and mouse anti-human CD56 (Clone NCAM-1; PE conjugated, BD Pharmingen) were used to confirm monocyte and NK cell purity following enrichment and to determine monocyte and NK cell subsets. Plasma used for ADCC experiments were collected from 9 different HIV-infected individuals, heat-inactivated for 1h at 56°C and conserved as previously described17 until ready to use in subsequent experiments.
CD4-mimetic BNM-III-170
The small molecule CD4-mimetic BNM-III-170 synthesized as previously described69 was dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 10 mM, aliquoted, and stored at − 20°C prior to being diluted to 50μM in PBS for cell-surface staining and ADCC assays.
Viral stock production, infections, and detection of infected cells
Vesicular stomatitis virus G (VSVG)-pseudotyped viruses were produced in 293T by Polyethylenimine (PEI) transfection. Briefly 1 × 107 293T seeded in a TC75cm2 flasks (Cat# 734–2315; VWR) were transfected with plasmids encoding VSVG and HIV infectious molecular clone (IMC) at a ratio of 2:5. 4h post transfection, the supernatant was removed and replaced with fresh media and cultured for a further 72h. Cell supernatants were harvested, clarified by centrifugation, and filtered through a 0.45μM filter (Minisart) and layered onto a 25% sucrose gradient prior to ultracentrifugation. Pseudotyped viruses (concentrated 100-fold) were subsequently titrated on CD4+ T cells as described21. In addition day 7 differentiated macrophages were cultured in 3.5cm petri dishes and passively infected for 5 days to determine macrophage specific inoculum required to obtain >10% infection.
Flow cytometry analysis of cell-surface staining and ADCC responses
Five days post-infection, macrophages were washed in PBS, incubated in 10 mM EDTA for 30 min at RT, detached and transferred to 96-well V-bottom plates (Corning; Cat # 0877126). In parallel, CD4+ T cells infected for 48h were harvested, washed and transferred to 96-well V-bottom plates and incubated for 30 min with AquaVivid viability dye (Thermo Fisher Scientific, Cat# L43957) as per manufacturer’s instructions. Cells were then washed twice in PBS. Prior to staining with antibodies, macrophages were incubated with 10% human sera (Valley Biomedicals) and 2% FcBlock (Miltenyi) in FACS buffer (1% BSA, 1mM EDTA in PBS) for 10 min. Following Fc blocking, macrophages and CD4+ T cells resuspended in 1% BSA were incubated with a panel of anti-Env antibodies (Table S2; key resources table) pre-coupled to CF647 fluorophore (Sigma-Aldrich) for 30 min at RT. Binding of 19b, 17b, A32, F240 and 10E8 was done with or without BNM-III-170 (50μM). Cells were then washed twice with FACS buffer, fixed with 2% Paraformaldehyde (PFA) and permeabilized using BD CytoFix/CytoPerm Fixation/Permeabilization Kit (BD Biosciences) as per manufacturer’s instructions. Detection of p24 + infected cells was performed as described21,52 using anti-Gag p24-FITC (Beckman Coulter). The percentage of infected cells (p24 + cells) was determined by gating the living cell population based on the viability dye staining (Aqua Vivid, Thermo Fisher Scientific, Cat# L43957). Samples were analyzed on a Fortessa cytometer (BD Biosciences, Mississauga, ON, Canada) and data analysis was performed using Flow Jo version 10.4.0 (Tree Star, Ashland, OR, USA).
Measurement of ADCC-mediated killing was performed as previously described.17,22 Briefly, primary CD4+ T cells infected for 48h with the AD8 wild type (WT) virus or viruses defective for Nef and Vpu (N-U-) were co-cultured with autologous PBMC (Effector: Target ratio of 10:1), autologous NK (Effector: Target ratio of 1:1) or autologous monocytes (Effector: Target ratio of 2:1) in the presence of HIV + plasma (1:1000) with or without BNM-III-170 or bNAbs. Macrophage ADCC assays were performed as above with the following modifications; macrophages infected for 5 days with AD8 WT or N-U- were incubated with 10% human sera prior to co-culture with effectors and HIV + plasma (1:1000) with or without BNM-III-170 or bNAbs for 5h at 37°C. The percentage of cytotoxicity was calculated as described.17,22
QUANTIFICATION AND STATISTICAL ANALYSES
Statistics were analyzed using GraphPad Prism version 9.3.1 (GraphPad, San Diego, CA, USA). All datasets with statistical analysis was tested for statistical normality and this information was used to apply the appropriate statistical tests. p values <0.05 were considered significant; significance values are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Circular edge bundling graphs were generated in undirected mode in program R v.4.1.278 using ggraph, igraph, tidyverse, and RColorBrewer packages. Edges are only shown if p < 0.05, and nodes are sized according to the sum of the connecting edges’ absolute r values. Nodes are color-coded according to the groups of variables. Correlograms were generated using the corrplot and RColorBrewer packages using hierarchical clustering based on the principal component. Normalized heatmaps with dendrograms were created using the complexheatmap and tidyverse packages. Normalizations were done per column (analysis).
Supplementary Material
Highlights.
Nef, Vpu, and Env are required to fully downregulate CD4 from infected CD4+ T cells
Any combination of two of these proteins are sufficient in infected macrophages
Infected macrophages are resistant to ADCC mediated by HIV+ plasma
Small CD4 mimetics sensitize infected macrophages to ADCC mediated by HIV+ plasma
ACKNOWLEDGMENTS
The authors thank the CRCHUM BSL3 and Flow Cytometry Platforms for technical assistance, Mario Legault from the FRQS AIDS, and Infectious Diseases network for cohort coordination and clinical samples. We thank Dennis Burton (The Scripps Research Institute) for the JR-FL infectious molecular clone and Michel Nussenzweig for 3BNC117 and 101074 antibodies. This study was supported by grants from the National Institutes of Health to A.F. and J.C.K. (R01 AI148379), to A.F. (R01 AI129769 and R01 AI150322), to B.H.H. (R01 AI162646 and UM1 AI164570), and from the Basic Science Core of the University of Alabama at Birmingham Center for AIDS Research (AI27767). This work was also partially supported by 1UM1AI164562-01, co-funded by National Heart, Lung, and Blood Institute, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, National Institute on Drug Abuse and the National Institute of Allergy and Infectious Diseases, a CIHR foundation grant #352417, a CIHR Team grant #422148, and a Canada Foundation for Innovation grant #41027 to A.F. A.F. is the recipient of a Canada Research Chair on Retroviral Entry #RCHS0235 950-232424. F.K. is supported by the Deutsche Forschungsgemeinschaft (CRC 1279 and SPP 1923). A.L. and R.G. were supported by a MITACS Accélération postdoctoral fellowship. A.L. was supported by a ViiV Healthcare postdoctoral fellowship during the revision of the manuscript. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111983.
REFERENCES
- 1.von Bredow B, Arias JF, Heyer LN, Moldt B, Le K, Robinson JE, Zolla-Pazner S, Burton DR, and Evans DT (2016). Comparison of antibody-dependent cell-mediated cytotoxicity and virus neutralization by HIV-1 env-specific monoclonal antibodies. J. Virol. 90, 6127–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand SP, Ding S, Tolbert WD, Prévost J, Richard J, Gil HM, Gendron-Lepage G, Cheung WF, Wang H, Pastora R, et al. (2021). Enhanced ability of plant-derived PGT121 glycovariants to eliminate HIV-1-Infected cells. J. Virol. 95, e0079621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richard J, Prévost J, Baxter AE, von Bredow B, Ding S, Medjahed H, Delgado GG, Brassard N, Stürzel CM, Kirchhoff F, et al. (2018). Uninfected bystander cells impact the measurement of HIV-specific antibody-dependent cellular cytotoxicity responses. mBio 9, 003588–e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruel T, Guivel-Benhassine F, Amraoui S, Malbec M, Richard L, Bourdic K, Donahue DA, Lorin V, Casartelli N, Noël N, et al. (2016). Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat. Commun. 7, 10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton KL, Mylvaganam G, Villasmil-Ocando A, Stuart H, Maus MV, Rashidian M, Ploegh HL, and Walker BD (2021). HIV-infected macrophages resist efficient NK cell-mediated killing while preserving inflammatory cytokine responses. Cell Host Microbe 29, 435–447.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian C, Yun Z, Yao Y, Cao M, Liu Q, Hu S, Zhang S, and Luo D (2019). Heterogeneous macrophages: supersensors of exogenous inducing factors. Scand. J. Immunol. 90, e12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginhoux F, and Guilliams M (2016). Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449. [DOI] [PubMed] [Google Scholar]
- 8.Wallet C, De Rovere M, Van Assche J, Daouad F, De Wit S, Gautier V, Mallon PWG, Marcello A, Van Lint C, Rohr O, and Schwartz C (2019). Microglial cells: the main HIV-1 reservoir in the brain. Front. Cell. Infect. Microbiol. 9, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borrajo A, Spuch C, Penedo MA, Olivares JM, and Agís-Balboa RC (2021). Important role of microglia in HIV-1 associated neurocognitive disorders and the molecular pathways implicated in its pathogenesis. Ann. Med. 53, 43–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cenker JJ, Stultz RD, and McDonald D (2017). Brain microglial cells are highly susceptible to HIV-1 infection and spread. AIDS Res. Hum. Retroviruses 33, 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho DD, Rota TR, and Hirsch MS (1986). Infection of monocyte/macrophages by human T lymphotropic virus type III. J. Clin. Invest. 77, 1712–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson JK, Cross GD, Callaway CS, and McDougal JS (1986). In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV). J. Immunol. 137, 323–329. [PubMed] [Google Scholar]
- 13.Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, and Burke DS (1988). Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167, 1428–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jambo KC, Banda DH, Kankwatira AM, Sukumar N, Allain TJ, Heyderman RS, Russell DG, and Mwandumba HC (2014). Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol. 7, 1116–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B, Sharron M, Montaner LJ, Weissman D, and Doms RW (1999). Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 96, 5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veillette M, Désormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, et al. (2014). Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J. Virol. 88, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prévost J, Richard J, Gasser R, Medjahed H, Kirchhoff F, Hahn BH, Kappes JC, Ochsenbauer C, Duerr R, and Finzi A (2022). Detection of the HIV-1 accessory proteins nef and vpu by Flow Cytometry represents a new tool to study their functional interplay within a single infected CD4+ T cell. J. Virol. 96. e0192921–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prévost J, Richard J, Medjahed H, Alexander A, Jones J, Kappes JC, Ochsenbauer C, and Finzi A (2018). Incomplete downregulation of CD4 expression affects HIV-1 env conformation and antibody-dependent cellular cytotoxicity responses. J. Virol. 92. 004844–e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brand D, Srinivasan K, and Sodroski J (1995). Determinants of human immunodeficiency virus type 1 entry in the CDR2 loop of the CD4 glycoprotein. J. Virol. 69, 166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, and Hendrickson WA (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veillette M, Coutu M, Richard J, Batraville LA, Dagher O, Bernard N, Tremblay C, Kaufmann DE, Roger M, and Finzi A (2015). The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J. Virol. 89, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schön A, Freire E, Routy JP, et al. (2015). CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc. Natl. Acad. Sci. USA 112, E2687–E2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding S, Veillette M, Coutu M, Prévost J, Scharf L, Bjorkman PJ, Ferrari G, Robinson JE, Stürzel C, Hahn BH, et al. (2016). A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J. Virol. 90, 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prévost J, Zoubchenok D, Richard J, Veillette M, Pacheco B, Coutu M, Brassard N, Parsons MS, Ruxrungtham K, Bunupuradah T, et al. (2017). Influence of the envelope gp120 phe 43 cavity on HIV-1 sensitivity to antibody-dependent cell-mediated cytotoxicity responses. J. Virol. 91, 024522–e2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, and Weiss RA (1984). The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312, 763–767. [DOI] [PubMed] [Google Scholar]
- 26.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, and Montagnier L (1984). T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312, 767–768. [DOI] [PubMed] [Google Scholar]
- 27.Delwart EL, and Panganiban AT (1989). Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J. Virol. 63, 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson RE, Sanfridson A, Ottinger JS, Doyle C, and Cullen BR (1993). Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J. Exp. Med. 177, 1561–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wildum S, Schindler M, Münch J, and Kirchhoff F (2006). Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J. Virol. 80, 8047–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BK, Gandhi RT, and Baltimore D (1996). CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 70, 6044–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alsahafi N, Ding S, Richard J, Markle T, Brassard N, Walker B, Lewis GK, Kaufmann DE, Brockman MA, and Finzi A (2015). Nef proteins from HIV-1 elite controllers are inefficient at preventing antibody-dependent cellular cytotoxicity. J. Virol. 90, 2993–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forthal DN, and Finzi A (2018). Antibody-dependent cellular cytotoxicity in HIV infection. AIDS 32, 2439–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Finzi A, and Sodroski J (2020). The conformational states of the HIV-1 envelope glycoproteins. Trends Microbiol. 28, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard J, Prévost J, Alsahafi N, Ding S, and Finzi A (2018). Impact of HIV-1 envelope conformation on ADCC responses. Trends Microbiol. 26, 253–265. [DOI] [PubMed] [Google Scholar]
- 35.Laumaea A, Smith AB 3rd, Sodroski J, and Finzi A (2020). Opening the HIV envelope: potential of CD4 mimics as multifunctional HIV entry inhibitors. Curr. Opin. HIV AIDS 15, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arias JF, Heyer LN, von Bredow B, Weisgrau KL, Moldt B, Burton DR, Rakasz EG, and Evans DT (2014). Tetherin antagonism by Vpu protects HIV-infected cells from antibody-dependent cell-mediated cytotoxicity. Proc. Natl. Acad. Sci. USA 111, 6425–6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, and Chen BK (2014). HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J. Virol. 88, 6031–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giese S, Lawrence SP, Mazzon M, Nijmeijer BM, and Marsh M (2020). The nef protein of the macrophage tropic HIV-1 strain AD8 counteracts human BST-2/tetherin. Viruses 12, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard J, Prévost J, von Bredow B, Ding S, Brassard N, Medjahed H, Coutu M, Melillo B, Bibollet-Ruche F, Hahn BH, et al. (2017). BST-2 expression modulates small CD4-mimetic sensitization of HIV-1-Infected cells to antibody-dependent cellular cytotoxicity. J. Virol. 91, 002199–e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. (2011). Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. (2009). Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doores KJ, and Burton DR (2010). Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84, 10510–10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kek H, Laumaea A, Parise S, Poumbourios P, Hearps AC, and Jaworowski A (2021). Differential expression of HIV envelope epitopes on the surface of HIV-Infected macrophages and CD4(+) T cells. Antiviral Res. 191, 105085. [DOI] [PubMed] [Google Scholar]
- 44.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. (2010). Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. (2013). Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. (2014). Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. (2012). Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, and Blumenthal R (2007). Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 46, 1398–1401. [DOI] [PubMed] [Google Scholar]
- 49.Gohain N, Tolbert WD, Orlandi C, Richard J, Ding S, Chen X, Bonsor DA, Sundberg EJ, Lu W, Ray K, et al. (2016). Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240. Sci. Rep. 6, 36685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding S, Tolbert WD, Prévost J, Pacheco B, Coutu M, Debbeche O, Xiang SH, Pazgier M, and Finzi A (2016). A highly conserved gp120 inner domain residue modulates env conformation and trimer stability. J. Virol. 90, 8395–8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tolbert WD, Gohain N, Veillette M, Chapleau JP, Orlandi C, Visciano ML, Ebadi M, DeVico AL, Fouts TR, Finzi A, et al. (2016). Paring down HIV env: design and crystal structure of a stabilized inner domain of HIV-1 gp120 displaying a major ADCC target of the A32 region. Structure 24, 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richard J, Veillette M, Ding S, Zoubchenok D, Alsahafi N, Coutu M, Brassard N, Park J, Courter JR, Melillo B, et al. (2016). Small CD4 mimetics prevent HIV-1 uninfected bystander CD4 + T cell killing mediated by antibody-dependent cell-mediated cytotoxicity. EBioMedicine 3, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prévost J, Tolbert WD, et al. (2019). An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25, 578–587.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madani N, Princiotto AM, Mach L, Ding S, Prevost J, Richard J, Hora B, Sutherland L, Zhao CA, Conn BP, et al. (2018). A CD4-mimetic compound enhances vaccine efficacy against stringent immunodeficiency virus challenge. Nat. Commun. 9, 2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anand SP, Prévost J, Baril S, Richard J, Medjahed H, Chapleau JP, Tolbert WD, Kirk S, Smith AB 3rd, Wines BD, et al. (2019). Two families of env antibodies efficiently engage fc-gamma receptors and eliminate HIV-1-Infected cells. J. Virol. 93, 018233–e1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajashekar JK, Richard J, Beloor J, Prévost J, Anand SP, Beaudoin-Bussières G, Shan L, Herndler-Brandstetter D, Gendron-Lepage G, Medjahed H, et al. (2021). Modulating HIV-1 envelope glycoprotein conformation to decrease the HIV-1 reservoir. Cell Host Microbe 29, 904–916.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw GM, Levy PC, and LoBuglio AF (1978). Human monocyte antibody-dependent cell-mediated cytotoxicity to tumor cells. J. Clin. Invest. 62, 1172–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohl S, Starr SE, oleske JM, Shore SL, Ashman RB, and Nahmias AJ (1977). Human monocyte-macrophage-mediated antibody-dependent cytotoxicity to herpes simplex virus-infected cells. J. Immunol. 118, 729–735. [PubMed] [Google Scholar]
- 59.Beaudoin-Bussières G, Arduini A, Bourassa C, Medjahed H, Gendron-Lepage G, Richard J, Pan Q, Wang Z, Liang C, and Finzi A (2022). SARS-CoV-2 accessory protein ORF8 decreases antibody-dependent cellular cytotoxicity. Viruses 14, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kramski M, Schorcht A, Johnston APR, Lichtfuss GF, Jegaskanda S, De Rose R, Stratov I, Kelleher AD, French MA, Center RJ, et al. (2012). Role of monocytes in mediating HIV-specific antibody-dependent cellular cytotoxicity. J. Immunol. Methods 384, 51–61. [DOI] [PubMed] [Google Scholar]
- 61.Ding S, Gasser R, Gendron-Lepage G, Medjahed H, Tolbert WD, Sodroski J, Pazgier M, and Finzi A (2019). CD4 incorporation into HIV-1 viral particles exposes envelope epitopes recognized by CD4-induced antibodies. J. Virol. 93, e01403–e01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. (2011). Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, et al. (2012). Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA 109, E3268–E3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. (2011). Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cavacini LA, Emes CL, Wisnewski AV, Power J, Lewis G, Montefiori D, and Posner MR (1998). Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res. Hum. Retroviruses 14, 1271–1280. [DOI] [PubMed] [Google Scholar]
- 66.Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, and Peden KW (1996). Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12, 191–194. [DOI] [PubMed] [Google Scholar]
- 67.Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, and Hahn BH (1991). Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65, 3973–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, et al. (2012). Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 86, 2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Melillo B, Liang S, Park J, Schön A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, et al. (2016). Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med. Chem. Lett. 7, 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Stürzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, et al. (2016). Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19, 504–514. [DOI] [PubMed] [Google Scholar]
- 71.Ding S, Grenier MC, Tolbert WD, Vézina D, Sherburn R, Richard J, Prévost J, Chapleau JP, Gendron-Lepage G, Medjahed H, et al. (2019). A new family of small-molecule CD4-mimetic compounds contacts highly conserved aspartic acid 368 of HIV-1 gp120 and mediates antibody-dependent cellular cytotoxicity. J. Virol. 93, 013255-19-e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kmiec D, Iyer SS, Stürzel CM, Sauter D, Hahn BH, and Kirchhoff F (2016). Vpu-mediated counteraction of tetherin is a major determinant of HIV-1 interferon resistance. mBio 7, 009344–e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emi N, Friedmann T, and Yee J-K (1991). Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J. Virol. 65, 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prévost J, Tolbert WD, Medjahed H, Sherburn RT, Madani N, Zoubchenok D, Gendron-Lepage G, Gaffney AE, Grenier MC, Kirk S, et al. (2020). The HIV-1 env gp120 inner domain shapes the Phe43 cavity and the CD4 binding site. mBio 11, 002800–e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bar KJ, Tsao C-y, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, et al. (2012). Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 8, e1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, et al. (2013). Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Stürzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, et al. (2016). The potency of nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.R Core Team. R (2017). A Language and Environment for Statistical Computing (R Foundation for Statistical Computing; ). [cited 2022 5th June]. Available from: https://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.


