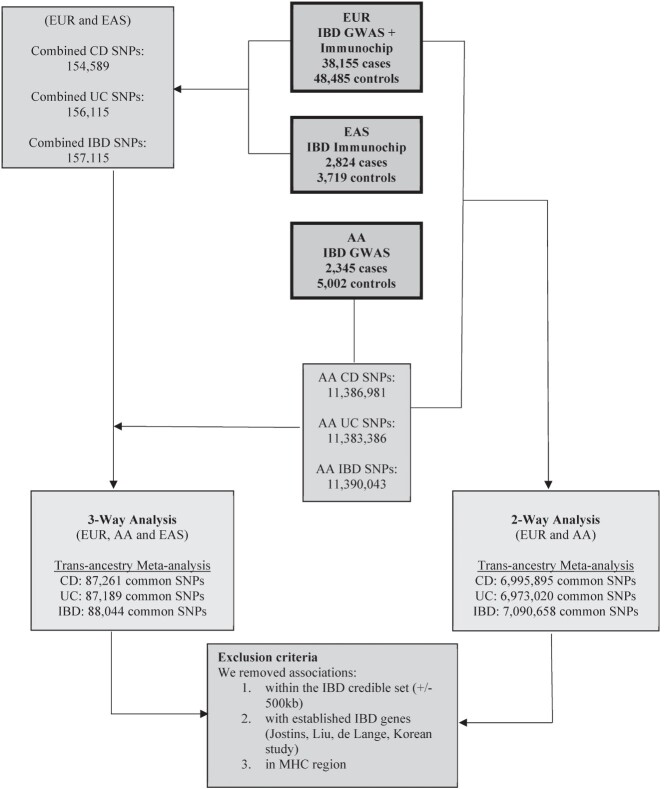
Figure 2.
Workflow to identify novel SNP associations with CD, UC and IBD. This is an overview of the workflow used in determining new risk loci in the IBD trans-ancestry meta-analysis. Summary statistics from EUR, EA and AA cohorts were combined using MANTRA for each of the phenotypes of interest (UC, CD and IBD). The left-hand side of the figure depicts the common SNPs carried over in the downstream analysis for the EUR, EAS and AA trans-ancestry meta-analysis. The right-hand side of the figure displays the common SNPs carried over in the downstream analysis for the EUR and AA trans-ancestry meta-analysis. SNPs with a log 10 Bayes Factor ≥ 6 were considered to be genome-wide significant. Variants within 500 Kb of established risk loci for the IBD phenotype of interest or in the MHC region were removed from the analysis.