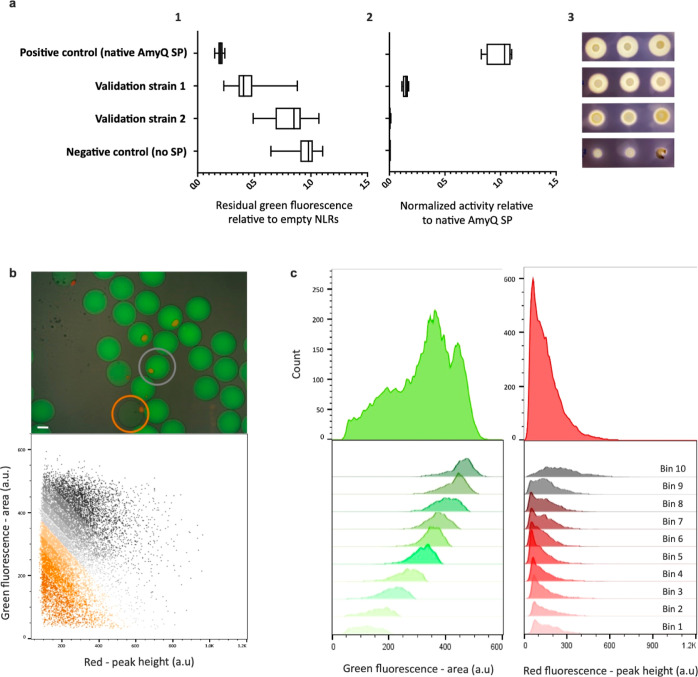
Figure 2.
HT SP library screening in NLRs. (a) For initial validation of the NLR-based α-amylase assay, four B. subtilis strains secreting AmyQ to different levels were used: three strains with known SP amino acid sequences at the N-terminus (one of them with the native SP of AmyQ; positive control) and a B. subtilis strain synthesizing the amylase without an N-terminal SP (negative control). Amylase secretion of each strain was assessed using the (1) NLR-based assay, (2) MTP colorimetric assay, and (3) starch hydrolysis test. (1) For the NLR-based assay, the values represent the residual fluorescein-labeled starch still present in the occupied NLRs after cell growth, relative to the green fluorescence of the empty NLRs in the same population (set as 1). The recorded events were positive control, 24 occupied and 850 empty NLRs; validation strain 1, 99 and 4329; validation strain 2, 50 and 932; negative control, 124 and 859. (2) For the MTP assay, the values are calculated relative to the amylase activity produced by the positive control (having a value of 1) and four biological replicates were performed. (3) Starch hydrolysis tests based on the starch–iodine reaction.23 (b) Top: overlay of bright-field and fluorescence microscopy images of NLRs after incubation in medium. Empty NLRs (no red dot) show a homogenous green fluorescence profile (no starch degradation), while NLRs harboring a colony (red dot) show different degrees of fluorescein-labeled starch degradation (orange circle: high secreter; gray circle: low secreter). Scale bar: 200 μm. Bottom: dot plot representing all occupied NLRs from one experiment (approximately 20,000 NLRs). The gating applied during the second sorting step is depicted in orange-gray color codes, which defines bins with distinct AmyQ secretion levels. (c) Green and red fluorescence profiles of all sorted events from the same experiment, both as a whole population (i.e., occupied NLRs; top panel) and divided into 10 equally sized bins (lower panel).