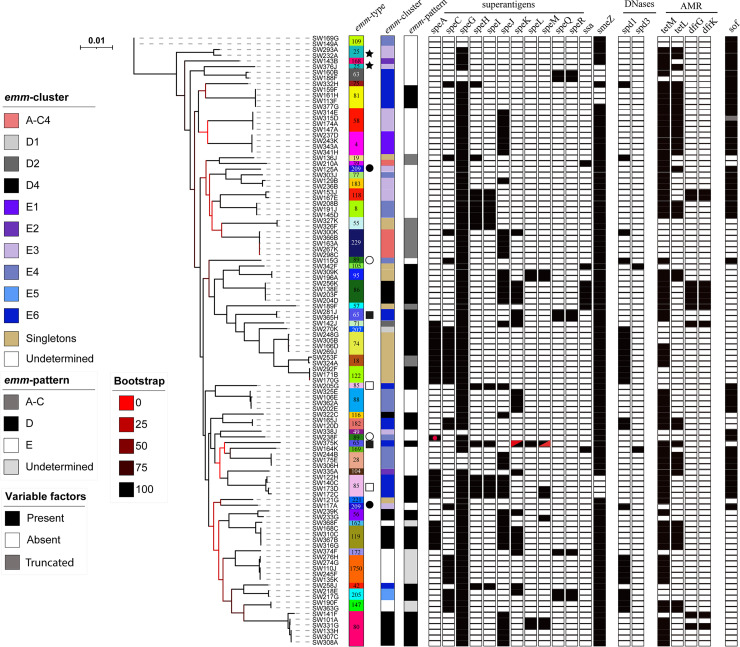
FIG 2.
Phylogenetic analysis of the 107 Gambian LIC isolate genomes. A maximum likelihood phylogeny was constructed from the core gene alignment (1,382,412 bp) using RAxML (51) with 100 bootstraps. Isolates clustered by emm type except for those indicated, whereby two lineages were represented by a single emm genotype: star, emm25; filled square, emm65; open square, emm85; open circle, emm89; filled circle, emm209. Also shown is the presence (black)/absence (white) of the superantigen genes speA, speC, speG, speH to speM, speQ, speR, ssa, and smeZ and DNase genes spd1 and spd3; four other DNase genes (sda1, sda2, sdn, and spd4) were tested for but were not found in any isolate. In all cases except one (red dot), speA was located within the prophage-like element Φ10394.2. One isolate had a gene that appeared to be a fusion of 5′ speK and 3′ speM (red triangles). Antimicrobial resistance genes (AMR) tetM, tetL, dfrG, and dfrK were also identified in some isolates (white, absent; black, present). The positivity for serum opacity factor (sof) is also shown, although for one emm55 isolate, this gene would produce a truncated variant of serum opacity factor (gray). The scale bar represents substitutions per site. emm types are colored for easy visualization, and type numbers are also given.