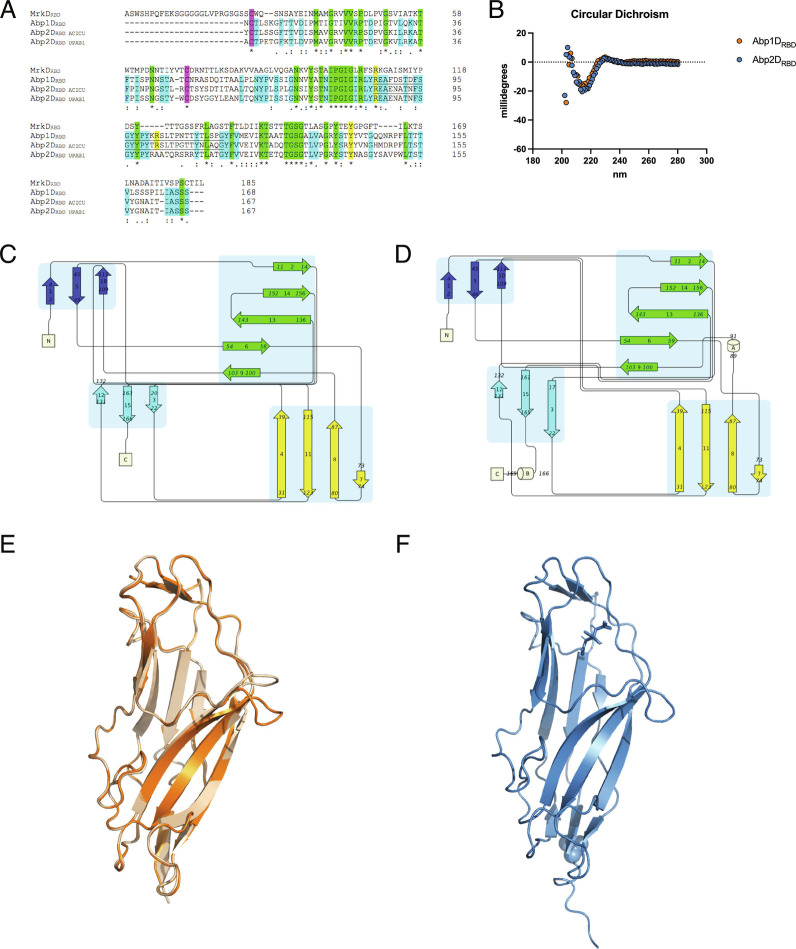
Fig. 3.
Structural features of Abp1DRBD and Abp2DRBD. (A) Receptor binding domain alignment of MrkD (3U4K), Abp1D, Abp2DACICU, and Abp2DUPAB1. Alignment by Clustal Omega. Purple—disulfide bond cysteines. Green—amino acid identical to all four amino acid sequences. Blue—residues identical to the three Acinetobacter sequences. Yellow—residues of interest in RBD binding mechanism. (B) Circular dichroism spectrum of Abp1DRBD and Abp2DRBD in 20 mM Phosphate 7.00. Portion of spectrum shown is as indicated by the axes here and as in the supplemental materials. (C and D) Topology maps of Abp1DRBD and Abp2DRBD (Pro-Origami). (E and F) Crystal structures of Abp1DRBD (molecule A in orange and molecule B in wheat, rmsd 0.176 Å) (8DF0) and Abp2DRBD (blue) (8DEZ) (PyMol).