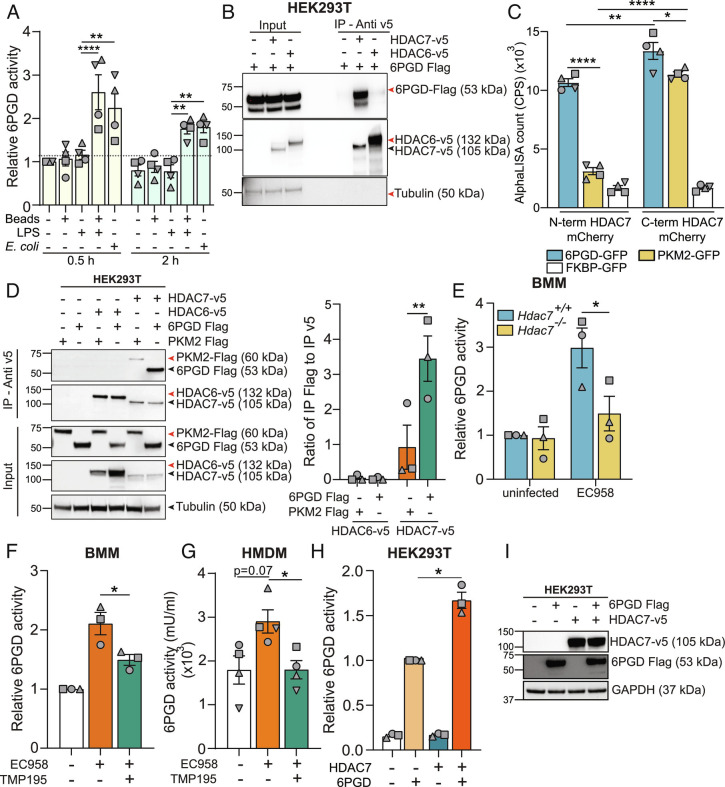
Fig. 5.
HDAC7 interacts with and activates 6PGD. (A) 6PGD activity assay on lysates from BMM that had been treated for 0.5 h or 2 h with either LPS (1 ng/mL), latex beads, or LPS plus latex beads or infected with EC958 (MOI 100). (B) Immunoblot of coimmunoprecipitation assay using HEK293T lysates to assess 6PGD interactions (representative of three independent experiments). (C) AlphaLISA assay to analyze the interaction between HDAC7 and 6PGD, PKM2, or FKBP (negative control) following cell-free expression of indicated proteins. (D) Immunoblot (representative of three independent experiments, Left) of coimmunoprecipitation assay using HEK293T lysates to assess strength of HDAC7 interaction with target proteins (6PGD versus PKM2). Interactions were quantified by densitometric analysis of FLAG-tagged proteins (Right). (E) 6PGD activity assay on lysates from the indicated BMM populations infected with EC958 for 2 h. (F and G) 6PGD activity assay on lysates from BMM (F) or HMDM (G) pretreated with TMP195 (10 μM) for 1 h followed by infection with EC958 for 2 h. (H and I) 6PGD activity assay on lysates from HEK293T cells cotransfected with the indicated constructs (H). Immunoblotting was performed on the lysates to confirm similar expression of the proteins (I, representative of three independent experiments). Graphical data (mean ± SEM, n = 3 to 4) are combined from at least three independent experiments and are normalized to untreated control (A), Hdac7+/+ uninfected control (E), uninfected control (F), or 6PGD alone (H). 6PGD enzyme activity was normalized to the total protein in the lysate (A, E–G). Statistical significance was determined using two-way ANOVA (A and C–E), repeated measure one-way ANOVA with Giesser Greenhouse correction (F and H), or nonparametric Kruskal–Wallis test (G) followed by Tukey’s, Dunn’s, Dunnett’s, or Sidak’s multiple comparison test (ns, not significant; *P < 0.05; **P < 0.01; ****P < 0.0001).