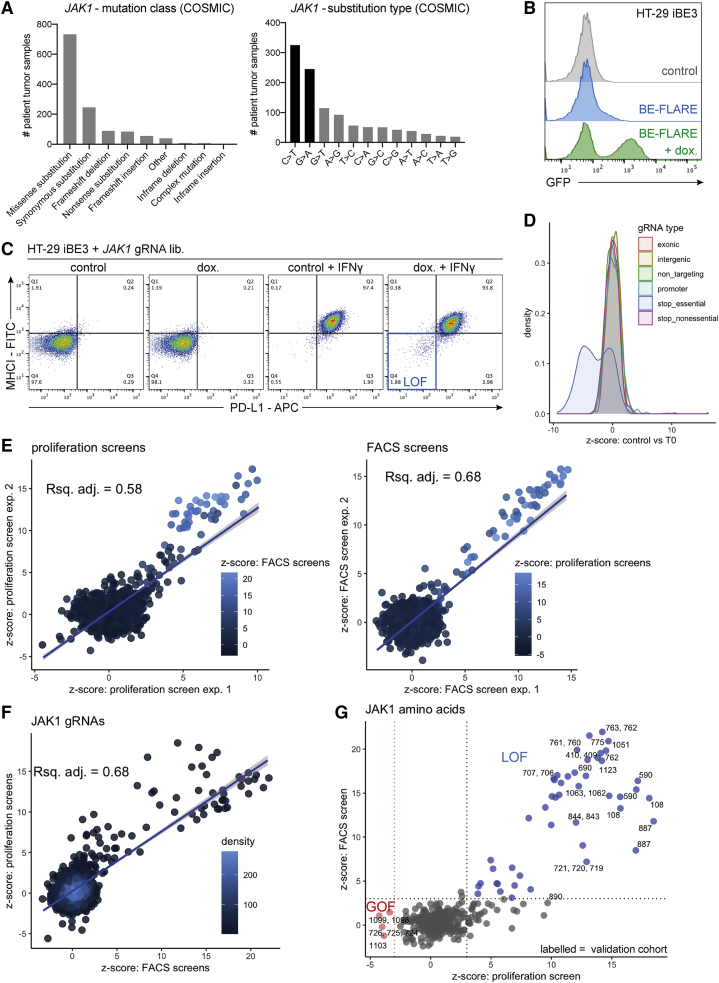
Figure 2.
Base editing mutagenesis screening of JAK1 variants
(A) COSMIC data from patient tumor samples show JAK1 cancer mutations are predominantly C->T and G->A missense variants.
(B) BE-FLARE assessment of base editing efficiency in HT-29 iBE3 cells treated with doxycycline, based on flow cytometry analysis of a BFP (His66) to GFP (Tyr66) spectral shift.
(C) FACS screening assay. After base editing of JAK1 by the addition of doxycycline, HT-29 iBE3 cells that failed to respond to IFN-γ after 48 h were selected by FACS, as determined by the lack of induction of MHC-I and PD-L1 expression.
(D) Proliferation screening assay. gRNA depletion or enrichment is indicated by z-score, comparing the control arm with the T0 (time 0) control. Base editing gRNAs designed to introduce stop codons in essential genes in HT-29 iBE3 cells are depleted.
(E) Correlation between screening replicates. z-scores for gRNAs targeting JAK1 were compared between replicates and alternative screening assays. The shaded line represents the 95% confidence interval.
(F) Correlation between different base editor screening assays for JAK1 variants in HT-29 iBE3 cells.
(G) Identification of LOF and GOF alleles in JAK1 protein affecting sensitivity to IFN-γ. z-scores for the base editing screens using FACS vs proliferation were plotted to select potential LOF (blue) and GOF (red) JAK1 variants. Labeling illustrates amino acid positions that were selected for further validation. All data are representative of two independent experiments or screens performed on separate days. See also Figure S2 and Table S2.