Figure 6.
Verification of base editing genotypes with next-generation sequencing
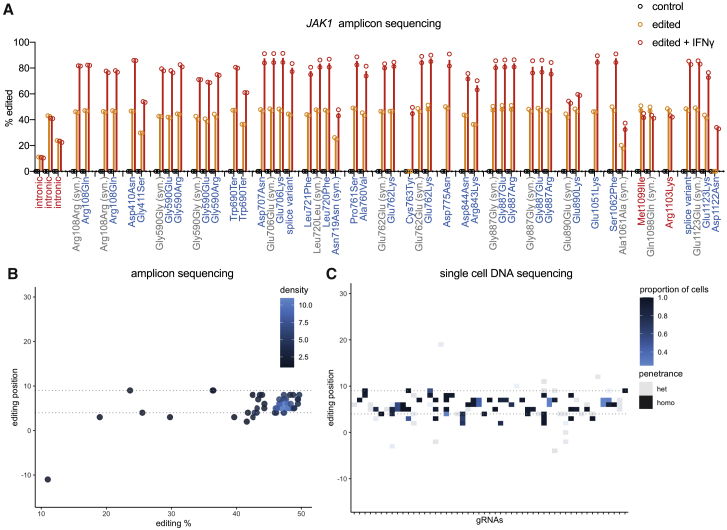
(A) Deep sequencing of JAK1 reveals the DNA editing profile of base editor gRNAs. Editing variant allele frequency for predicted LOF and GOF gRNAs within the validation cohort measured by NGS of amplicons in control cells, base edited cells, or base edited cells with selection with IFN-γ for 6 d. Different editing outcomes are grouped by gRNA. Syn, synonymous. Data represent the mean of two independent experiments performed on separate days.
(B) Amplicon sequencing assessment of HT-29 iBE3 base editing positions within the gRNA protospacers profiled in Figure 6A in the absence of IFN-γ.
(C) Single-cell DNA sequencing of base editing in HT-29 iBE3 cells across 50 gRNAs reveals C->T (or G->A) editing focused in the gRNA activity window. Penetrance (zygosity) 0/1/1 is heterozygous (het), and 1/1/1 is homozygous (homo). The proportion of cells with the same gRNA assignment harboring that edit is indicated. See also Figure S6 and Table S4.