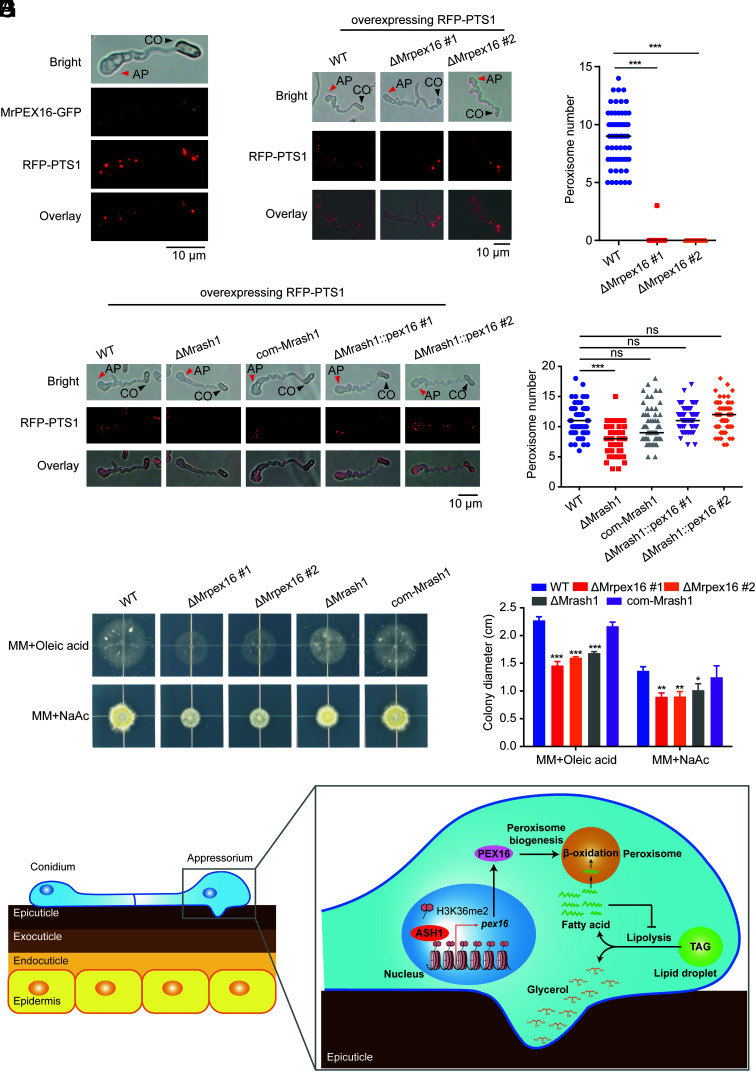
Fig. 5.
MrASH1 activates Mrpex16 to regulate peroxisome biogenesis and fatty acid utilization in M. robertsii. (A) Subcellular locations of PEX16 fused with GFP and peroxisomes marked with RFP–PTS1 in the appressorium of WT induced in MM-Gly medium on hydrophobic plates for 20 h. CO, conidium; AP, appressorium. (B) Subcellular locations of peroxisomes marked with RFP–PTS1 in the appressoria of the WT and two ΔMrpex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. CO, conidium; AP, appressorium. (C) Quantification of peroxisomes in the appressoria of the WT and two ΔMrpex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. In total, 50 to 100 appressoria were detected for each strain. Horizontal lines represent the medians. Significant differences in the median compared with those in WT were determined by Student's t test. ***P < 0.001. The experiments were repeated twice with similar results. (D) Subcellular locations of peroxisomes marked with RFP–PTS1 in the appressoria of the WT, ΔMrash1, com-Mrash1, and two ΔMrash1::pex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. CO, conidium; AP, appressorium. (E) Quantification of peroxisomes in the appressoria of the WT, ΔMrash1, com-Mrash1, and two ΔMrash1::pex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. In total, 50 to 100 appressoria were detected for each strain. Horizontal lines represent the medians. Significant differences in the median compared with those in WT were determined by Student's t test. ***P < 0.001. ns, not significant. The experiments were repeated twice with similar results. (F) Fungal colonies of the WT, two ΔMrpex16 strains, ΔMrash1, and com-Mrash1 strains grown on MM plates supplied with 2.5 mM oleic acid (a long-chain fatty acid) or 50 mM NaAc (sodium acetate, a short-chain fatty acid) for 7 d. (G) Colony diameters of the WT, two ΔMrpex16 strains, ΔMrash1, and com-Mrash1 strains on fatty acid or acetate plates. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. *P < 0.05, **P < 0.01, and ***P < 0.001. The experiments were repeated twice with similar results. (H) Schematic model showing the regulatory mechanism of the ASH1–PEX16 regulatory pathway in appressorium turgor generation and cuticle penetration in the insect pathogenic fungus. Upon host cuticle exposure, the up-regulated epigenetic regulator MrASH1 activates the target gene Mrpex16 via H3K36me2 modification on chromatin to control peroxisome biogenesis in the appressorium. Efficient degradation of fatty acids in the peroxisome via β-oxidation prevents their accumulation from inhibiting lipolysis and in turn promotes the hydrolysis of TAGs within lipid droplets to produce large amounts of glycerol for appressorium turgor pressure generation.