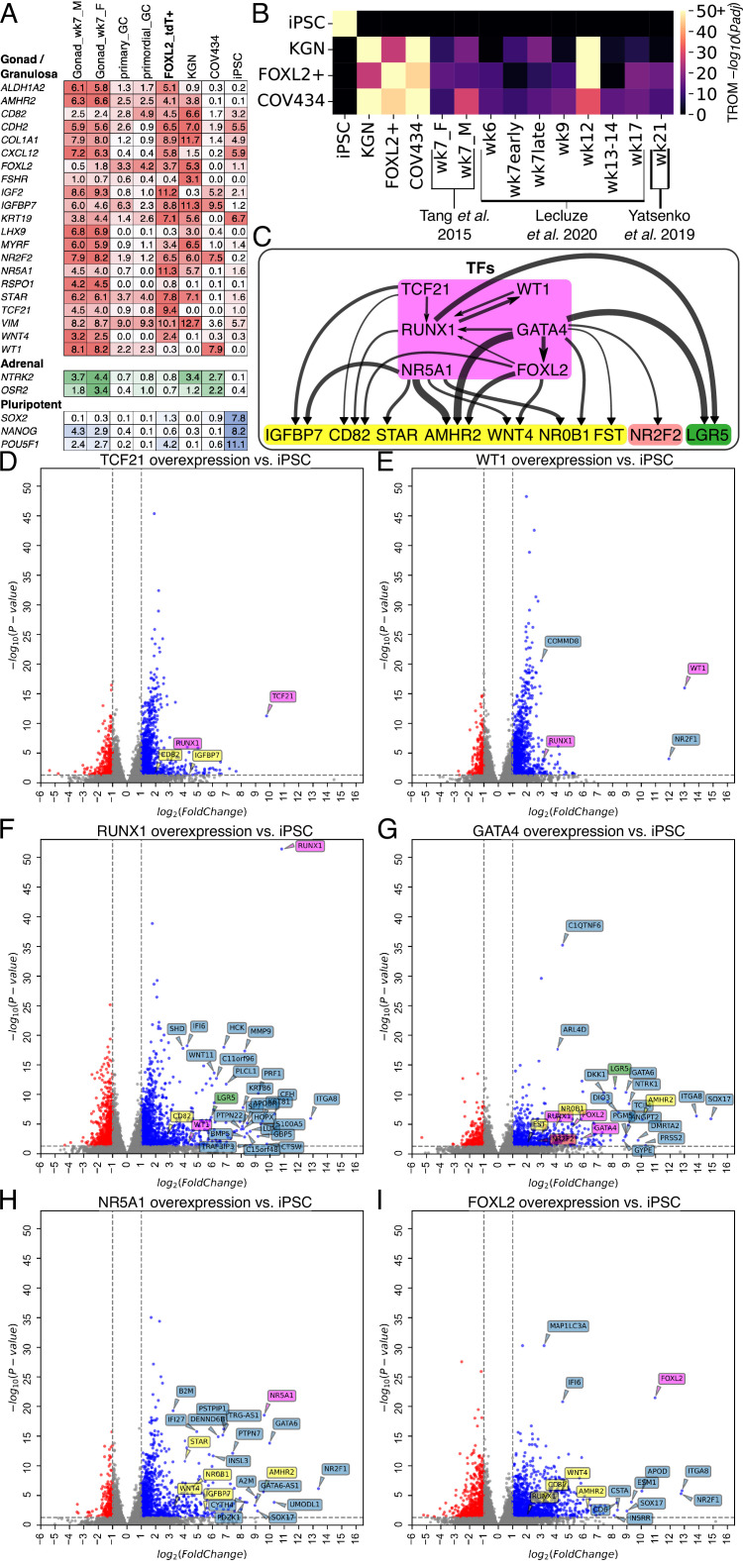
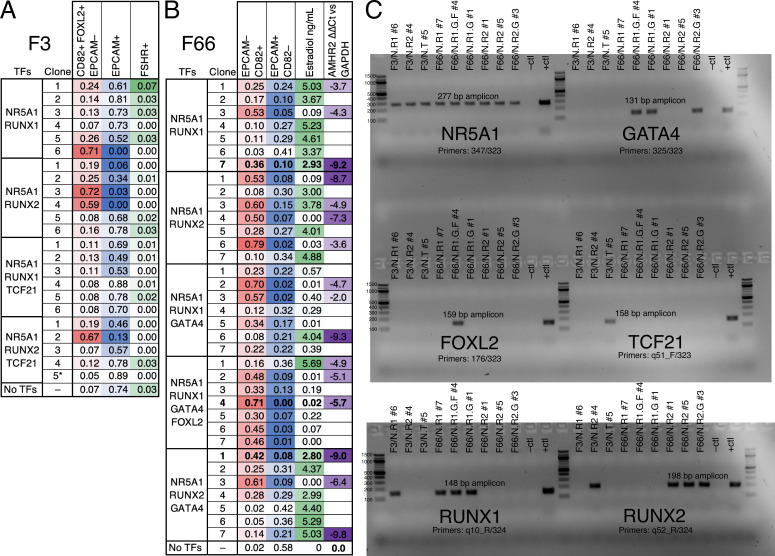
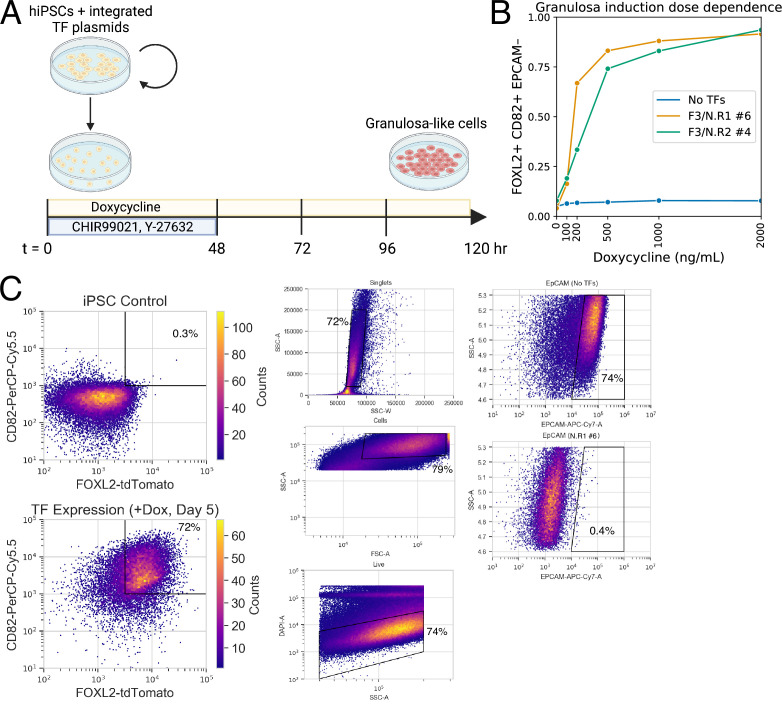
Figure 3. Transcriptomic analysis of transcription factor (TF)-induced granulosa-like cells.
(A) Gene expression of selected markers in granulosa-like cells. Log2(TPM) values for gondal/granulosa, adrenal, and pluripotent marker genes were compared between 7 wpf male and female fetal gonad somatic cells, primary and primordial granulosa cells, TF-induced FOXL2+ cells, KGN cells, COV434 cells, and human induced pluripotent stem cells (hiPSCs). (B) Transcriptome overlap measure (TROM) comparison of TF-induced FOXL2+ cells, COV434 cells, and hiPSCs with published in vivo data from different time points in ovarian development. (C) Regulatory effects of granulosa-related TFs. RNA-seq was performed after 2 days of TF overexpression in hiPSCs (TFs shown in magenta). A differential gene expression (DEG) analysis was performed for all samples relative to the hiPSC control (n = 2 biological replicates each). Black arrows represent significant (false discovery rate <0.05) upregulation, with the width proportional to the log2-fold change. Interactions are shown between TFs (magenta) and granulosa marker genes (yellow), as well as the stromal/theca marker NR2F2 (red) and the pre-granulosa marker LGR5 (green). (D–I) Volcano plots showing DEGs in the TF overexpression experiments. Colors are as in panel C; other DEGs not listed in panel C are shown in blue. Not all DEGs could be labeled due to space limits, but they are listed in the Source Data for this figure.