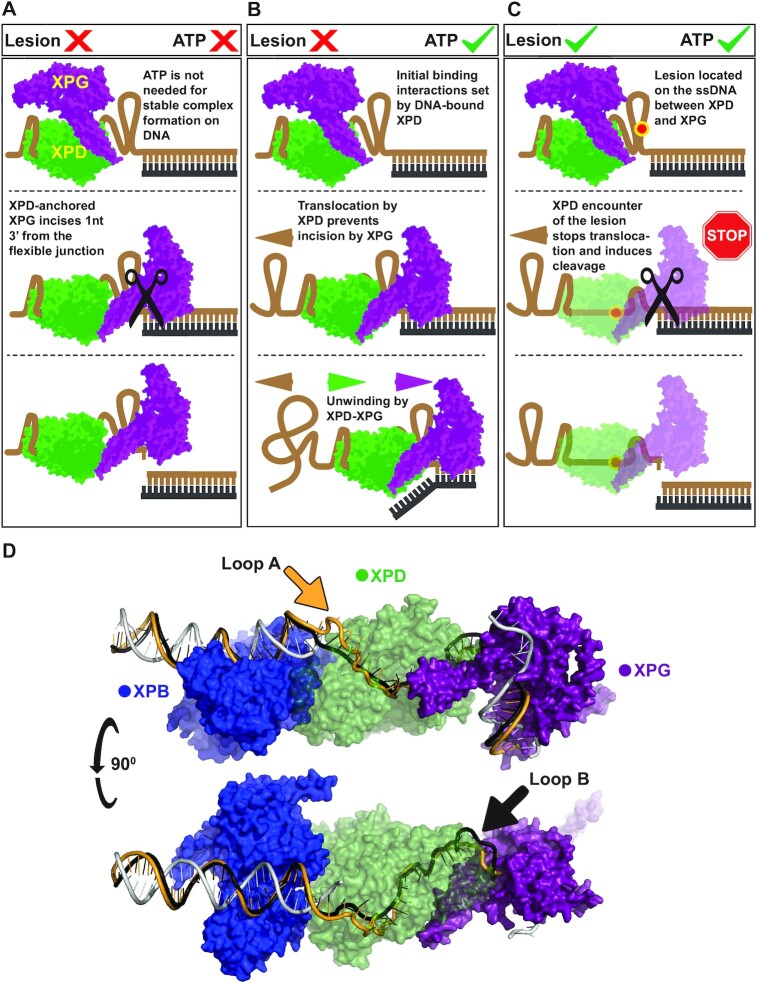
Figure 5.
Structural models that rationalize coreTFIIH-XPG activity on the ss/ds junction based on existing structural information and our experimental results and analyses. Initial binding to the substrate proceeds via the 5′ end by coreTFIIH which has a higher affinity for the substrate compared to XPG. For simplicity, the XPD domain of coreTFIIH alone is represented (green) along with XPG (purple). XPD recruits XPG via DNA-mediated protein-protein interactions. The ssDNA between XPD and XPG is highly condensed or extruded to enable direct XPD-XPG interactions even if XPG is located at the duplex. (A) The absence of ATP precludes translocation which enables access and flexibility at the junction, resulting in incision. (B) In the presence of ATP, DNA is pulled through XPD by the action of ATP hydrolysis. The ATP-hydrolyzing/translocating XPD prevents incision by XPG. Once all of the ssDNA is translocated, the complex begins unwinding which proceeds to completion in the absence of a lesion. (C) In the presence of a lesion, the XPD-XPG complex translocates until stopped by the lesion. This stalling and the potential disruption of ATP-hydrolysis trigger XPG incision. (D) A model of XPB (blue), XPD (green), and XPG (purple) interactions at the opened bubble substrate. The non-translocating strand is shown when bound by XPB, and the rest is omitted as it would require Replication protein A (RPA) docking. The translocating strand is shown in gold and black. The two paths show two possibilities of looping DNA out (arrows), to enable efficient DNA handling and XPD-XPG interaction. The model showcases the sharp bending of the 3’ duplex by XPG which enables active site positioning while maintaining XPD-XPG contacts.