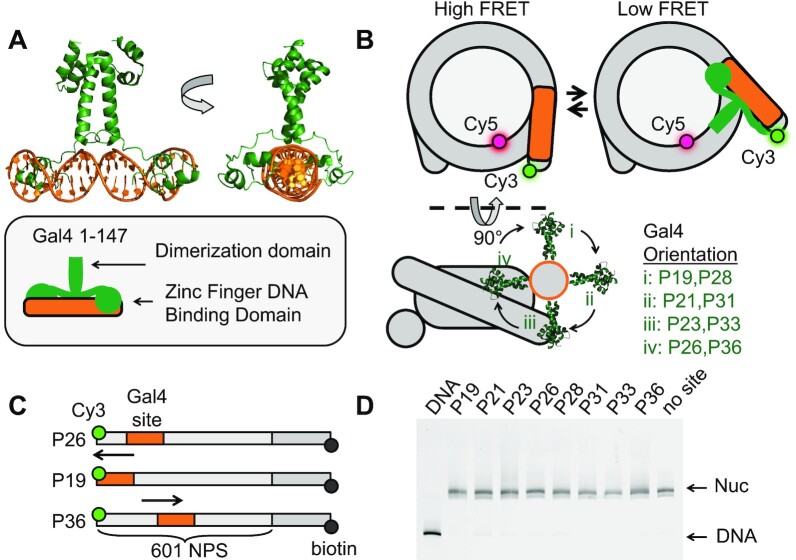
Figure 1.
Experimental design to probe Gal4 binding throughout nucleosome entry-exit region. (A) Crystal structure of the transcription factor (TF) Gal4 bound to DNA as a dimer (PDB 3COQ) (28). The side view shows how Gal4 encompasses 270 degrees of DNA when bound. In this study, we are using amino acids 1–147 which includes the DNA binding domain and the dimerization domain. (B) Simplified cartoon of the nucleosome containing a FRET pair between Cy5-H2A(K119C) and Cy3-DNA. Gal4 binds by trapping the nucleosome in an unwrapped state which is detectable by FRET. Lower diagram illustrates the rotational position of Gal4 with respect to the octamer. This diagram illustrates how movement of the of the binding site by 5bp results in a 180-degree shift of the binding site with respect to the octamer. (C) Nomenclature used to refer to binding site position (i.e. P26 means binding site ends 26 bp from DNA edge). All binding sites were inserted into the Widom 601 nucleosome positioning sequence (NPS) (27). Additionally, the DNA contains a 75 bp linker terminated with a biotin required for surface tethering in single-molecule experiments. (D) All nucleosomes used in this study have the same mobility through a poly-acrylamide native gel indicating that insertion of the Gal4 binding site does not influence nucleosome positioning.