Figure 2.
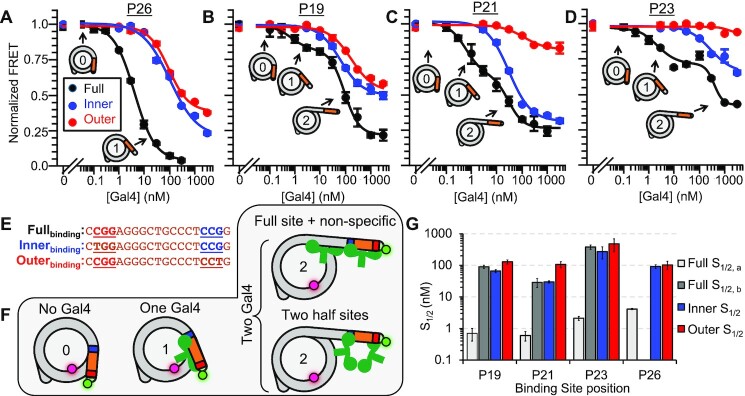
Gal4 binds with nanomolar affinity to Gal4 sites contained before the 30 bp barrier. (A–D) Ensemble FRET measurements of Gal4 binding to nucleosomes with a binding site at the position indicated above the plot. To the full binding site (black), the binding curves for P19, P21 and P23 exhibit two inflection points and are fit to the sum of two binding isotherms, while P26 is fit to a single binding isotherm. To the half site mutants, the FRET decrease begins at significantly higher concentrations and is fit to only one binding isotherm. Error bars indicate ±1 SEM. (E) Gal4 binding sites used in this study. In the ‘inner’ (blue) and ‘outer’ (red) DNA sequences, a ‘T’ is swapped into the 3 bp region recognized by the Zn-finger. This DNA mutation weakens the interaction of a Gal4 monomer with DNA. (F) A model describing multiple states of nucleosome unwrapping. State 1 represents Gal4 bound to its specific site within a nucleosome. This specific binding facilitates additional Gal4 binding and, subsequently, further nucleosome unwrapping. (G) Summary of S1/2 values from all fits. Error bars indicate ±1 standard error of the mean (SEM).