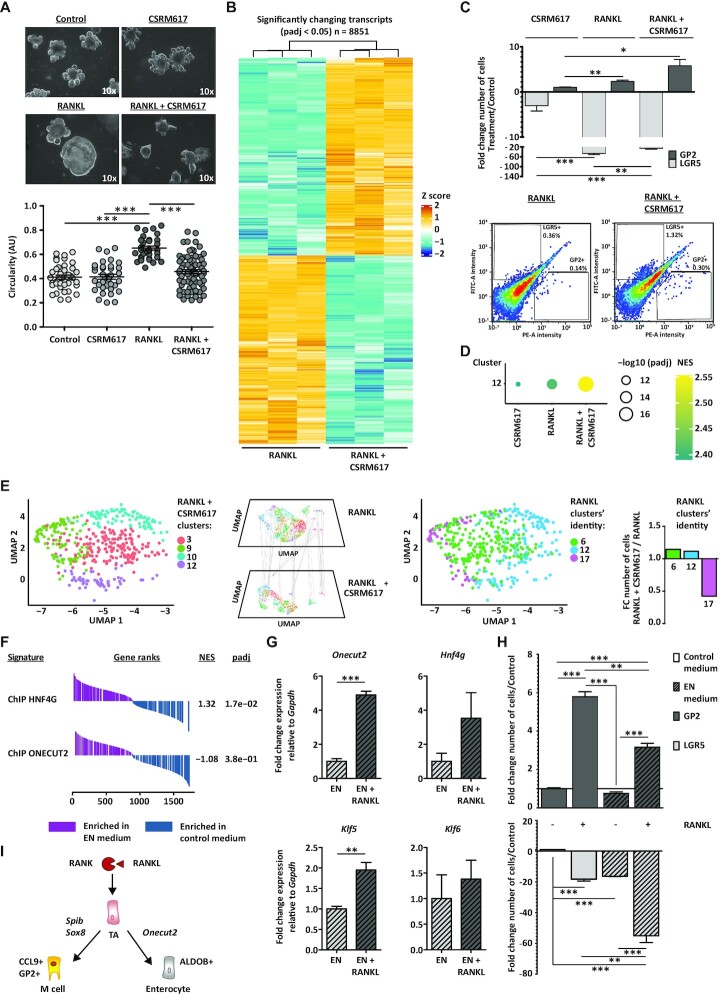
Figure 4.
ONECUT2 restricts M cell lineage specification in vitro. (A) Bright-field images of control organoids or those treated with RANKL, ONECUT2 inhibitor CSRM617 or a combination of both. Microscope magnification is depicted. Below, organoid circularity quantification graph (AU, arbitrary units) and mean ± SEM. Control n = 40, CSRM617 n = 35, RANKL n = 33 and RANKL + CSRM617 n = 70. (B) Heatmap showing the relative mRNA expression of transcripts upon co-treatment with RANKL and CSRM617, compared to RANKL alone. Rows show z scores of normalized, log2-transformed values from significantly changing genes (Padj < 0.05). (C) Bar graph representing the mean ± SEM of at least four differentiation experiments. Below, density plots depicting fluorescent intensities in FITC (LGR5) and PE (GP2) channels, determined by flow cytometry. Percentage of gated populations is described. (D) Gene-set enrichment analysis (GSEA) showing RANKL-treated cluster 12-specific gene signature, enriched in organoids treated as in (A), compared to control. Dotplot graph shows normalized enrichment score (NES, colour) and FDR (size). (E) Label transferring from RANKL-treated cell clusters within GOI1 to cells co-treated with RANKL and CSRM617. Bar graph (right) shows the fold change (FC) number of cells with RANKL clusters’ identity (RANKL + CSRM617 over RANKL). (F) GSEA showing the HNF4G- and ONECUT-ChIP signatures, enriched in organoids growing in EN medium, compared to control medium. NES and padj values are described. (G) Relative mRNA expression of Onecut2, Hnf4g, Klf5 and Klf6 detected in organoids growing on EN medium alone or with RANKL. (H) Ratio of LGR5+ or GP2+ cell numbers in organoids growing on EN medium alone or with RANKL or regular RANKL medium over control, determined by flow cytometry. Bar graph representing the mean ± SEM of three differentiation experiments. For (A), (C), (G) and (H), unpaired t test P <0.05 is represented by (*), P <0.01 by (**) and P <0.001 by (***). (I) RANK/RANKL signalling is essential to maintain a proper balance between M cell and enterocyte cell differentiation.