Figure 6.
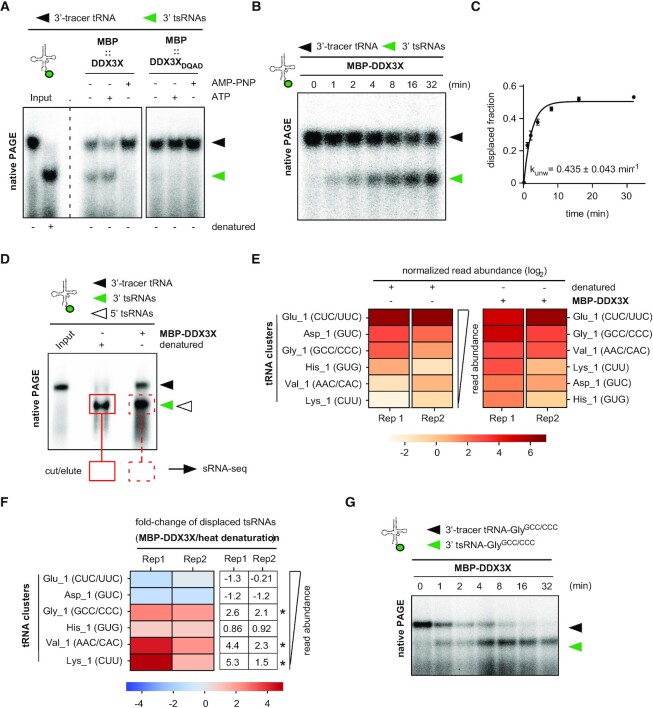
DDX3X separates 3′-tracer tRNAs with specificity for particular tRNA isoacceptors (A) Representative RNA helicase activity assay (fixed time-point) using equal molarities of recombinant MBP-DDX3X or MBP-DDX3XDQAD (750 nM) and 3′-tracer tRNAs (60 nM final) in the presence of equimolar ATP/MgCl2 or AMP-PNP/MgCl2 (2 mM). Black arrowhead, 3′-tracer tRNAs; green arrowhead, 3′ tsRNAs. (B) Representative RNA helicase activity assay using MBP-DDX3X (750 nM) and 3′-tracer tRNAs (20 nM final) in the presence of equimolar ATP/MgCl2 (2 mM). Aliquots were removed from reactions at indicated time points and separated using nPAGE. Arrowheads, as described in (A). (C) Quantification of triplicate time-course RNA helicase activity assays (see Supplemental Figure S5B) using MBP-DDX3X (750 nM) and 3′-tracer tRNAs (10 nM final) in the presence of equimolar ATP/MgCl2 (2 mM) to derive Kunw. Line marks the fit of the mean values to the integrated first-order rate equation, while error bars represent standard deviations. (D) Representative fixed-time point RNA helicase assay using MBP-DDX3X (750 nM) on 3′-tracer tRNAs (60 nM final) in the presence of equimolar ATP/MgCl2 (2 mM). 3′-tracer tRNAs that were heat-denatured were loaded in parallel. RNAs migrating at the level of heat-denatured and MBP-DDX3X-displaced 3′ tsRNAs (dashed and red squares, respectively) were excised in technical duplicates from nPAGE, cloned and subjected to small RNA sequencing. Black arrowhead, 3′-tracer tRNAs; green arrowhead, 3′ tsRNAs; white arrowhead, position where also non-labelled 5′ tsRNAs will migrate. (E) Heat maps representing relative read abundance (normalised log2 values) of known ANG tRNA substrates. The data was obtained by mapping replicate sRNA-seq data (columns) originating from heat-denatured 3′-tracer tRNAs (left panel) and MPB-DDX3X-displaced 3′-tsRNAs (right panel) to collapsed tRNA clusters as described in (52). Panels are ranked by relative abundance of the respective tRNA cluster in a descending manner. (F) Heat maps representing fold-changes of the relative read abundance (per tRNA cluster) after mapping reads originating from MPB-DDX3X-displaced tsRNAs divided by read abundance from heat-denatured 3′-tracer tRNAs (as described in (E)). Combined replicates (columns) ranked by relative abundance of the respective tRNA cluster in a descending manner. Asteriks denote tRNA isoacceptors with positive fold change ratios, indicating preferential activity of MBP-DDX3X on these tRNAs, which is independent of the corresponding tracer tRNA abundance in the 3′-tracer tRNA pool. (G) Representative time-course RNA helicase activity assay using MBP-DDX3X (750 nM) and 3′-tracer tRNA-GlyGCC/CCC (10 nM final) in the presence of equimolar ATP/MgCl2 (2 mM). Aliquots were removed from reactions at indicated time points and separated using nPAGE. Arrowheads, as described in (A).