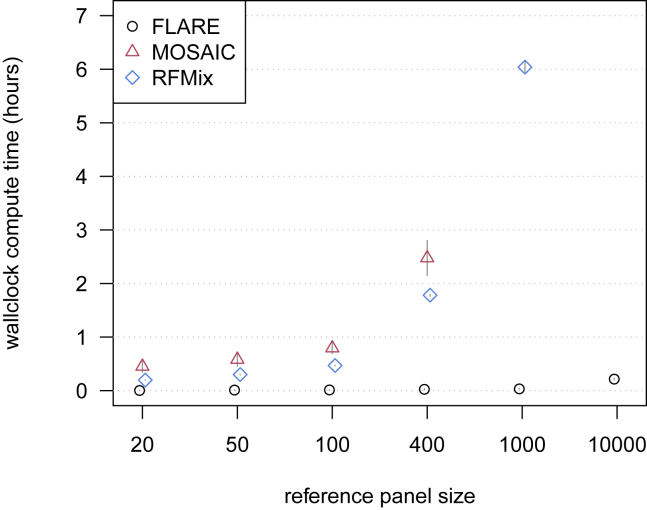
Figure 1.
Computation time for simulated sequence data with three-way admixture
Wallclock computation time in hours is shown on the y axis. Reference panel size for each of the three ancestries is shown on the x axis. Each analysis includes 100 admixed individuals, and results are averaged over four replicate simulations. Error bars (±2 standard errors) are shown as gray lines. Analyses of a simulated chromosome modeled on human chromosome 20 were run with 20 compute threads. MOSAIC could not analyze the data with 1,000 or more individuals per reference panel within the available 384 GB of computer memory. RFMix could not analyze the data with 10,000 individuals per reference panel within 48 h.