Abstract
Enterohemorrhagic Escherichia coli (EHEC) is an important gastrointestinal pathogen known for its ability to cause hemorrhagic colitis and induce hemolytic-uremic syndrome. The inner membrane QseC histidine kinase sensor has shown to be an important regulator of the locus of enterocyte effacement (LEE) island, where important EHEC key virulence genes are located. However, the QseC role during EHEC infection in human microbiota remains unknown. Herein, using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®), we investigated whether the QseC sensor has a role in human microbiota modulation by EHEC in a dynamic model. Our data demonstrated that the QseC sensor modulates human microbiota during EHEC infection, and its absence leads to an increase in Lactobacillaceae and Bifidobacterium genus predominance, although non-effect on Bacteroides genus by EHEC strains was observed. In co-culture, the Lactobacillus acidophilus has affected EHEC growth and impaired the EHEC growth under space-niche competition, although no growth difference was observed in the QseC sensor presence. Also, differences in EHEC growth were not detected in competition with Bacteroides thetaiotaomicron and EHEC strains did not affect B. thetaiotaomicron growth either. When investigating the mechanisms behind the SHIME results, we found that hcp-2 expression for the type 6 secretion system, known to be involved in bacterial competition, is under QseC sensor regulation beneath different environmental signals, such as glucose and butyrate. Our findings broaden the knowledge about the QseC sensor in modulating the human microbiota and its importance for EHEC pathogenesis.
Keywords: Enterohemorrhagic Escherichia coli, QseC, O157:H7, Human intestinal microbiota
Introduction
Enterohemorrhagic Escherichia coli (EHEC) is an important gastrointestinal pathogen known for its ability to cause hemorrhagic diarrhea, and due to its Shiga toxin production, it can lead to the development of uremic hemolytic syndrome (HUS) [1] that is known for inducing severe consequences in humans, such as kidney failure, being the leading cause of morbidity and mortality associated with outbreaks by this pathogen [2, 3]. Antibiotic treatments are not indicated for patients with EHEC infection since these drugs can induce through the SOS response, Shiga toxin production in EHEC, which is encoded by variants of the stx gene inserted into the EHEC chromosome via bacteriophage [1, 4]. Studying the microbiota to find probiotic competitors, as well as non-conventional antibiotic treatment, has emerged as a promising strategy to combat infections caused by EHEC.
EHEC has in its genome the Locus of Enterocyte Effacement pathogenicity island, which is activated by the Ler regulator for encoding proteins to the formation of the type III secretion system (T3SS) [5–7]. T3SS acts like a needle, injecting the Tir receptor into the host cell and binding to intimin, which is present in the bacterial cell membrane, thus promoting the close adhesion of the bacteria to the host cell [1]. The proteins inserted into the host promote changes in the cytoskeleton leading to the Attaching and Effacing (A/E) lesion [1, 7].
To detect changes in the environment and establish cell–cell communication, EHEC can recognize small chemical molecules or signals through receptors in their membrane [8, 9]. The membrane sensor histidine kinase QseC is a two-component system with its cognate response regulator QseB that responds to the adrenergic hormones norepinephrine and epinephrine of mammals and to the autoinducer-3 produced by bacteria [10]. Through the detection of changes in the environment by these signals, the QseC sensor regulates virulence genes in EHEC, such as the LEE pathogenicity island for the formation of A/E lesion in epithelial cells [11–14]. Although it is established that the sensor QseC is involved in EHEC pathogenicity, little is known about its contribution or regulation during EHEC interactions with the microbiota.
The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) is an in vitro model used in microbiology due to its combination of dynamism and multi-compatibility [15]. To date, a few studies have been conducted with EHEC in dynamic models [16–18]. SHIME® is a system composed of 5 bioreactors that include all compartments of the gastrointestinal tract, from the stomach to the colon, mimicking the same conditions of these compartments. In the bioreactors mimicking the colon, the model is kept under a fermentation process, in which the microbiota from donors is inoculated and stabilized to study the changes in the intestinal microbial community under different conditions [15, 19].
Understanding how EHEC behaves in different environments is essential to know the main mechanisms used by this microorganism in its adaptation to the host. In vivo and in vitro models have been used to explain EHEC infection mechanisms. Mice are important study models, but they do not have the same conditions as those found in human gastrointestinal (GI) microbiota. Also, EHEC infects murine, but they do not develop classic symptoms of the disease [5, 20]. In vitro evaluation in dynamic colonic models has been used to simulate the conditions of the human GI tract, to analyze the response of microorganisms in these compartments, and to better understand the complex community/dynamism of the human microbiota [15, 19]. Herein, we investigated whether the QseC sensor has a role in human microbiota modulation by EHEC in the SHIME dynamic model to better understand how different conditions and distinct metabolites via the QseC sensor may modulate the human microbiota and its importance for EHEC pathogenesis.
Methodology
Strains and culture conditions
All the strains used in this study are described in Table 1. The EHEC 8624 strains and their isogenic mutants were grown aerobically in Luria–Bertani (LB) broth supplemented with 50 Ug/ml streptomycin at 37 °C, overnight in a shaker (250 rpm). L. reuteri and L. acidophilus were grown in De Man, Rogosa, and Sharpe (MRS) broth, anaerobically at 37 °C, under static conditions.
Table 1.
Bacterial strains employed in the study
| Strains | Resistance Marker | Source |
|---|---|---|
| E. coli TOP10 with pBADMychisA(+ qseC) construction | Ampicillin | This study |
| Bacteroides thetaiotaomicron | - | [27] |
| Enterohemorrhagic E. coli 0157:H7 8624 strain (wild-type (WT)) | Streptomycin | [52] |
| 8624—mutant ΔqseC | Streptomycin | [53] |
| Lactobacillus acidophilus 3258, ATCC 4356 | - | André Tosello Foundation (http://fat.org.br) |
| Limosilactobacillus reuteri 3433, ATCC 23,272 | - | André Tosello Foundation (http://fat.org.br) |
Simulator of the Human Intestinal Microbial Ecosystem (SHIME®).
The test was carried out as described by Bianchi et al., 2018. SHIME® (registered trade name of the University of Ghent and ProDigest) is a simulator of the human intestinal microbial ecosystem that mimics conditions such as pH, residence time, and temperature through software [21, 22]. For the experiment, five double-coated reactors were used, one for simulating the stomach, one for the duodenum, and a triplicate for the ascending colon. The five reactors were continuously stirred with a magnetic stirrer, and the temperature was maintained at 37 °C. The system was maintained anaerobically through the daily introduction of N2 for 30 min. The colon pH (pH between 5.6 and 5.8) was automatically adjusted by adding 1 M NaOH or 0.1 M HCl [21, 22]. Each colon compartment received carbohydrate-based food that allows the adaptation of microorganisms to specific environmental conditions of the colon in terms of pH range, retention time, and available carbon sources, in volumes previously described. The stomach conditions and mimicking pancreatic juice (composed of Oxgall 6.0 g/L, NaHCO3 12.5 g/l, and pancreatin 0.9 g/l) were prepared as previously described [21, 22]. The colon reactors were inoculated with fecal microbiota samples from two donors, female and male healthy adults, ages between 20 and 26 years old, under a similar balanced diet and with no history of diarrhea or antibiotic use for at least 6 months prior to the trial, as previously described in the SHIME® usage [21, 22], and the absence of EHEC in the system was confirmed via the qRT-PCR assay prior to the infection. The feces were weighted, pooled, and diluted in 200 ml phosphate buffer containing Na2HPO4 0.05 mol/l, NaH2PO4 0.05 mol/l, and Na-thioglycolate 0.1% (pH 6.5); stirred for 10 min in a homogenizer (Stirrer model 130, Norte Científica, São Paulo, BR.); and centrifuged at 3000 × g for 15 min. The supernatants were subsequently added (40 ml) to the three colon reactors. The experimental protocol included a control period of 2 weeks (without intervention) after inoculation of the supernatant in the three colon reactors to adapt the microbial community to the nutritional and physical–chemical conditions and stabilize the microbiota [21]. During this period, 200 ml of the SHIME® feed entered the system, and 200 ml were discarded from each column reactor twice a day for 2 weeks. Once stabilization was carried out, 109 per ml of CFUs from the EHEC WT or ∆qseC strains were introduced into the stomach-mimicking reactor and were distributed to the duodenum portion and added the pancreatic juice until they were introduced into the last colon-mimicking reactor. Prior to any infection, a wash-out period (period necessary for new microbiota stabilization and elimination of the WT strain) was carried out, in which the microbiota received 200 ml of SHIME® food twice a day for 1 week, and the PCR analysis was performed to confirm the elimination of the WT strain from the reactors, followed by inoculation of the ∆qseC strain, as an independent subsequent experiment. For both the WT and ∆qseC strains, samples of the stomach and duodenum were collected, and CFU counts were performed to determine the concentration of bacteria after dilution in the stomach contents and to perform the analysis of bacterial cell viability at the stomach pH and pancreatic juice after 1 h of exposure. Samples were collected before introduction into the system (day 0 or D0) and after 24 h of infection. Total RNA was extracted to analyze the gene expression of the WT and ΔqseC strains and determine the abundance of the intestinal microbiota. For this purpose, analyses were carried out on the phyla Firmicutes, Bacteroidetes, and γ-Proteobacteria and on the genera Lactobacillaceae, Bifidobacterium, Prevotella, and Bacteroides, in addition to the virulence genes ler and stx-2 of the EHEC WT and ΔqseC.
Real-time qPCR
All RNA extractions were performed with Trizol and RiboPure-Bacteria RNA isolation kit (Ambion-Life), followed by the qRT-PCR technique. The total RNA was obtained at a concentration of 50 ng/μl per sample of the tests performed. For each reaction of 20 μl, Master Mix SYBR®, Multi-scribe® reverse transcriptase (Thermo Fisher Scientific), and RNAse inhibitor (Thermo Fisher Scientific) were used, in addition to 100 ng of sample RNA. The qRT-PCR reverse transcriptase reaction was performed in biological triplicates and experimental duplicates. The reactions were normalized with the RNA polymerase subunit A (rpoA) as an endogenous control to analyze the expression of virulence genes. As an endogenous control to identify the members of interest in the microbiota, eub338, a universal gene for Eubacteria, was used. QuantStudio3 equipment (Thermo Fisher Scientific) was used to carry out the reactions. The results obtained by the qRT-PCR assay were analyzed by the comparative critical threshold (ΔΔCT), as previously described (Walters and Sperandio, 2006). Error bars represent the standard deviation of the CT values. All primers used in this study are listed in Table 2.
Table 2.
Oligonucleotides used in the study
| Target | Primer set sequence 5′–3′ | Source | |
|---|---|---|---|
| Forward | Reverse | ||
| stx-2a | ACCCCACCGGGCAGTT | GGTCAAAACGCGCCTGATA | [54] |
| espA | TCAGAATCGCAGCCTGAAAA | GAAGGATGAGGTGGTTAAGCT | [14] |
| ler | CGACCAGGTCTGCCCTTCT | GCGCGGAACTCATCGAAA | [14] |
| hcp-2 | GAACGTCAGGCAGTTTCCGT | GGCCACGCTATCTGGTGAAA | [30] |
| rpoA | GCGCTCATCTTCTTCCGAAT | CGCGGTCGTGGTTATGTG | [14] |
| Bacteroides (RNA 16S) | CGATGGATAGGGGTTCTGAGAGGA | GCTGGCACGGAGTTAGCCGA | [55] |
| Prevotella (RNA 16S) | CACCAAGGCGACGATCA | GGATAACGCCYGGACCT | [55] |
| Delta e Gamma proteobacteria (RNA 16S) | GCTAACGCATTAAGTRYCCCG | GCCATGCRGCACCTGTCT | [56] |
| Eubacteria (Eub – RNA 16S) | ACTCCTACGGGAGGCAGCAGT | ATTACCGCGGCTGCTGGC | [57] |
| Firmicutes (RNA 16S) | TGAAACTYAAAGGAATTGACG | ACCATGCACCACCTGTC | [56] |
| Bacteroidetes (RNA 16S) | CRAACAGGATTAGATACCCT | GGTAAGGTTCCTCGCGTAT | [35] |
| Bifidobacterium (RNA 16S) | TCGCGTC(C/T)GGTGTGAAAG | CCACATCCAGC(A/G)TCCAC′ | [58] |
|
Lactobacillus (Lactobacillaceae) (RNA 16S) |
AGCAGTAGGGAATCTTCCA | CACCGCTACACATGGAG | [58] |
In vitro analysis in high and low glucose in co-culture with B. thetaiotaomicron
The WT and isogenic mutant strains were grown overnight and inoculated in tubes in the proportion of 30:1 of high D-MEM medium (4.5 g/l) or low glucose (1.0 g/l) plus inoculum in the presence or absence of B. thetaiotaomicron. In co-cultures, the bacteria proportions used were 1:1 EHEC and B. thetaiotaomicron. The tests were performed in anaerobic and static conditions at 37 °C. The bacterial growth was measured via turbidity reading after 1 h, 2 h, 4 h, 6 h, and 8 h incubation, and growth curves were plotted after optical density measurement. The experiments were conducted in biological triplicates and experimental duplicates.
EHEC co-culture with Lactobacillus acidophilus or Limosilactobacillus reuteri
EHEC, L. acidophilus, and L. reuteri strains were grown overnight. After the measurement of the optical densities of the cultures, the strains were inoculated in LB + MRS broth at a ratio of 1:1 or 1:5 of EHEC strains and L. acidophilus or L. reuteri. In the single culture group, the strains were grown alone. The strains were kept in an incubator on static interaction at 37 °C for 8 h, then the samples were diluted, plated on LB containing streptomycin for EHEC and MRS agar for L. acidophilus or L. reuteri, and the CFUs were counted. The experiments were conducted in biological triplicates and experimental duplicates.
Culture under sodium butyrate and glucose-mediated conditions
Bacterial cultures were grown in D-MEM low glucose or DMEM low glucose supplemented with But or NaCl until O.D. 1.0. Total RNA was extracted using trizol together with the RiboPure-Bacteria RNA isolation kit (Ambion-Life). After RNA extraction, gene expression analyses of virulence genes were performed. The experiments were conducted in biological triplicates and experimental duplicates.
Agar competition assay between EHEC and L. acidophilus or L. reuteri
The strains WT and ∆qseC and L. acidophilus and L. reuteri were grown overnight in LB and MRS media, respectively. The optical densities of the inoculum at 600 nm were measured. The lowest O.D. presented by the strains after 16 h of growth was used in the test. After adjusting the O.D., cultures were centrifuged at 7000 rpm for 2 min and resuspended in 300 μl PBS 1X. After this step, the ratio of 1:1 or 1:5 of the EHEC strains plus L. acidophilus or L. reuteri, respectively, was added in a new Eppendorf tube, centrifuged at 7000 rpm for 2 min, and resuspended in PBS 1X and 10 μl of the co-culture were inoculated on an LB + MRS (1:1) agar plates. In the single culture group, the strains were grown alone. After 16 h of interaction, the agar plates co-cultures were collected in tubes to perform serial dilutions and CFU counting on LB containing streptomycin for EHEC and MRS agar as a selective medium for L. acidophilus or L. reuteri (Peng et al., 2015). The experiments were conducted in biological triplicates and experimental duplicates.
Statistical analysis
The data were analyzed in the GraphPad Prism 8, and the statistical significance was determined by one-way analysis of variance (ANOVA). P values ≤ 0.05 were considered statistically significant.
Results
QseC modulates the intestinal fitness and microbiota shift promoted by EHEC infection
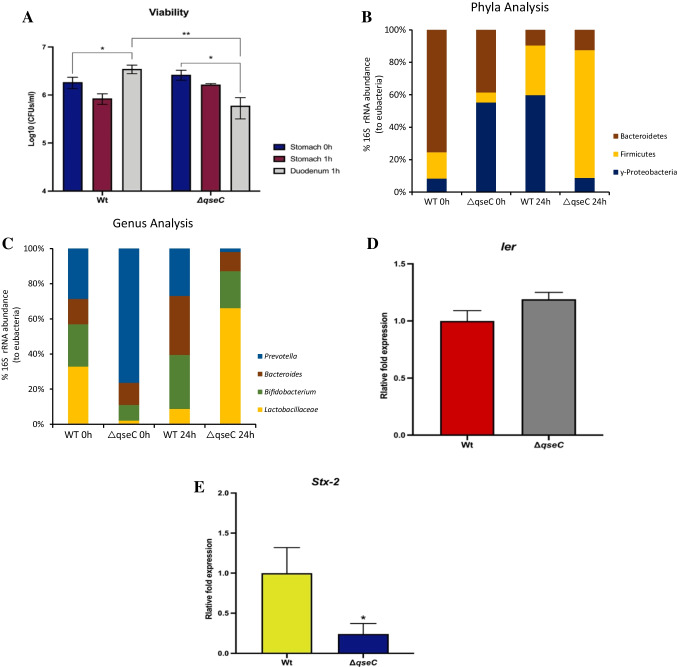
The sensor kinase QseC is an important bacterial communication sensor that helps EHEC to sense the environment and respond properly to its changes, modulating virulence genes accordingly with the niche inserted [23]. To evaluate whether the QseC sensor is involved in microbiota modulation by EHEC, we carried out an assay in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) to evaluate the direct competition between the EHEC WT and ΔqseC strains with the human microbiota. EHEC absence was confirmed via stx-2 and ler PCR prior to the infection (data not shown). The SHIME infection by EHEC has not been described in the literature, being an interesting and unprecedented work. Bacterial viability was controlled in the stomach and duodenum reactors. The strains were inoculated at the same rate in the stomach reactor at 0 h on different days (Fig. 1A). At 1 h in the stomach, both WT and ΔqseC have shown high resistance to low pH (pH 2.5). Nevertheless, after 1 h in the duodenum, the WT has recovered its levels, whereas the ΔqseC has presented a 22% CFU reduction when compared with its initial CFUs at stomach 0 h.
Fig. 1.
Microbiota and gene expression analyses from samples collected at SHIME before and after 24 h of EHEC WT and ΔqseC strain infection. A Analysis of the survival profile of EHEC WT and ΔqseC strains at SHIME. B The abundance of 16S rRNA to detect phyla ɣ-Proteobacteria, Bacteroidetes, Firmicutes, and C Lactobacillaceae, Bifidobacterium, Bacteroides, and Prevotella genus frequency was investigated via total RNA extraction for the RT-PCR assay analysis, 0 and 24 h after infection for the analysis. D and E The virulence genes expression analyses of ler and stx-2 samples collected from SHIME after 24 h of EHEC strains infection were performed. Statistical significance compared to the wild strain (WT). Bars without an asterisk show no statistically significant difference, p < 0.05 (*), p < 0.01 (**)
Next, to further evaluate how the microbiota could be affected by the strains at phyla and genera levels, microbiota members were determined in reactors mimicking the ascending colon, which is one of the predicted niches for the initial EHEC infection [6, 24]. The samples were collected after 24 h of interaction between EHEC strains and the microbiota at SHIME®. This period was chosen to allow EHEC colonization during SHIME® infection, once these bacteria in vivo depend on the attachment and effacement to survive and colonize the gut. Upon WT strain infection in the SHIME® model, almost 60% of the phyla analyzed belong to γ-Proteobacteria, which includes the inoculated EHEC and other Enterobacteriaceae members. The Firmicutes and Bacteroidetes phyla are essential members of the human microbiota; nevertheless, after 24-h infection, the proportion was 30% and 10%, respectively, of the total microbiota evaluated. Unlike the WT strain, during the ΔqseC mutant infection, there was a significant increase in the phylum Firmicutes, around 60% of the composition, followed by Bacteroidetes and γ-Proteobacteria, which together composed only 20% of the microbiota, when compared to 0-h microbiota composition (Fig. 1B). To analyze at genera level, microbiota members known for their beneficial performance in the intestine were chosen, such as Lactobacilliacea and Bifidobacterium genus, and members that have been demonstrated to be increased in some inflammatory diseases such as Prevotella and Bacteroides [25, 26]. The WT strain infection led to a Prevotella and Bacteroides ratio of 60% of the microbiota analyzed. On the other hand, the Lactobacillaceae and Bifidobacterium genera corresponded only 40% of the composition. Interestingly, for the ΔqseC mutant, there was an increase in approximately 70% Lactobacillaceae proportion, followed by 20% of Bifidobacterium, whereas Prevotella and Bacteroides were only 10% of the microbiota composition analyzed, when compared to 0-h microbiota composition (Fig. 1C).
We have also evaluated the virulence gene expression of Shiga toxin (stx-2) and LEE island (ler) genes. There was a significant decrease in the stx-2 gene for the ΔqseC strain compared with WT (Fig. 1D). On the other hand, ler, the master regulator of LEE island, had similar expression levels in the WT and ΔqseC strain (Fig. 1E). This result corroborates with the function of ler in EHEC, once this gene is important to perform A/E lesion to the intestinal wall, absent in this SHIME® model.
B. thetaiotaomicron does not impair EHEC growth under co-culture conditions
Previous studies indicated that B. thetaiotaomicron did not affect EHEC growth [27], and during SHIME® infection, there were no significant differences in the Bacteroides genus in the WT and ΔqseC strains. That way, we performed a co-culture assay to evaluate if EHEC and B. thetaiotaomicron has a direct impairment in EHEC growth. Also, the growth curve was performed under low and high-glucose conditions to evaluate whether the availability of carbon sources in different concentrations could trigger competition between the bacterial strains. The growth curves performed confirmed that B. thetaiotaomicron does not affect EHEC growth, even when the availability of sugar was decreased (Fig. 2).
Fig. 2.
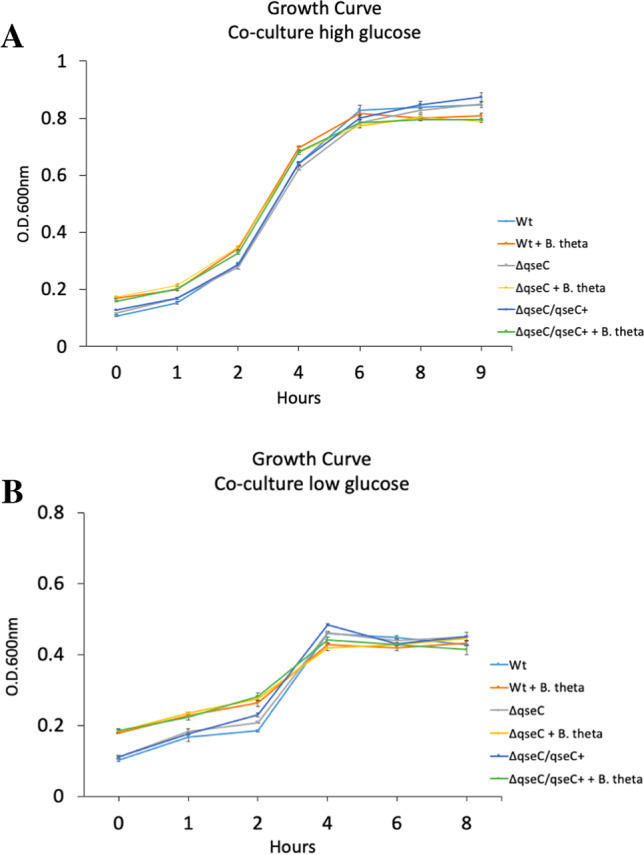
Growth curve of EHEC strains in high or low glucose in the presence or absence of B. thetaiotaomicron. The WT, ΔqseC, and ΔqseC/qseC + strains were cultured anaerobically in DMEM high-glucose (A) or low-glucose (B) medium in the presence or absence of Bacteroides thetaiotaomicron (B. theta) at 37 °C for 8 h. The cultures were kept under the static condition at 37 °C, and each time interval of 1, 2, 3, 4, 6, and 8 h aliquots were taken to read the optical density (O.D. 600 nm). Bars without an asterisk showed no statistically significant difference
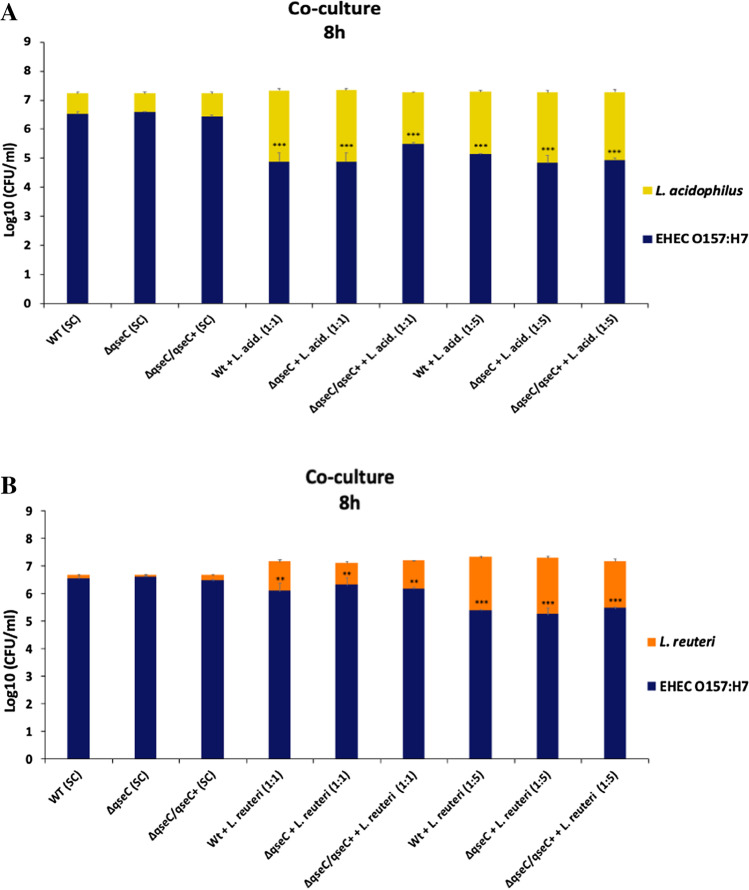
Lactobacillus acidophilus directly affects EHEC growth under co-culture conditions
During the SHIME® infection, the most prominent genus modulated by the QseC absence was Lactobacillaceae; we next carried out co-culture assays to analyze whether EHEC could affect Lactobacilli growth under QseC sensor regulation. Two different species of Lactobacillaceae were employed, Lactobacillus acidophilus 3258 (LA 3258) and Limosilactobacillus reuteri 3334 (LR 3334). Also, since the Lactobacillaceae is found in large amounts in the gut, two different proportions of Lactobacillaceae were evaluated: same proportion of EHEC and Lactobacillaceae (1:1) and fivefold Lactobacillaceae in comparison to EHEC (1:5). Different from the expected, L. acidophilus in both conditions (1:1 or 1:5) did not show a significant growth decrease when compared to its respective control group in a single culture. Conversely, L. acidophilus in co-culture with EHEC in both, 1:1 and 1:5, ratio caused a significant reduction in the EHEC growth when compared to their control group in the WT and ΔqseC strains single culture (Fig. 3A). Since at SHIME® there is a complex bacterial community, the WT and ΔqseC strains were also tested with Limosilactobacillus reuteri to evaluate whether we would observe a similar phenotype with Lactobacillaceae species. L. reuteri and EHEC strains demonstrated a similar growth profile in a single culture. Nevertheless, the 1:1 proportion led to a decrease in the EHEC growth in both WT and ΔqseC strains. In co-culture, in a 1:5 ratio with L. reuteri, EHEC strains’ growth inhibition was higher (Fig. 3B). These results indicate that L. reuteri and L. acidophilus could interfere in the EHEC growth, more pronounced in the presence of L. acidophilus; however, EHEC did not seem to affect the growth of these Lactobacillaceae species in co-culture conditions, and the QseC role here did not seem evident.
Fig. 3.
Broth co-culture assay between (A) Lactobacillus acidophilus or (B) Limosilactobacillus reuteri and EHEC strains. UFCs count after 8 h of interaction. The strains WT, ΔqseC, ΔqseC/qseC + , and L. acidophilus or L. reuteri were grown as single culture (SC) in broth. The strains WT + L. acid. or L. reuteri and ΔqseC + L. acid. and ΔqseC/qseC + + L. acid or L. reuteri were grown in broth co-culture at the ratio of 1:1 or 1:5 of EHEC plus L. acidophilus or L. reuteri, respectively. Bars without an asterisk showed no statistically significant difference, p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****) (two-way ANOVA)
The absence of glucose led to an increased gene expression of T6SS, and the sensor QseC is involved in the regulation of hcp-2
The gut environment has a high number of microorganisms struggling for nutrient availability in a niche where all cohabitants have evolved to compete for distinct nutrient sources. Bacteria have different mechanisms and forms to adapt to distinct conditions such as limited- and abundant-carbon sources, type secretion systems, and the production of antibacterial molecules to kill potential niche competitors [28]. The type VI secretion system works among bacterial communities through the introduction of bacteriolytic effectors into cells, usually involved in the pathogenic mechanisms of various pathogens [29, 30]. Due to the differences in the microbiota induced by WT and ΔqseC strains at SHIME®, we next investigated whether EHEC T6SS via QseC sensor kinase regulation could be involved in the results found. Thus, the assay was carried out in high- (4.5 g/l) and low-glucose (1 g/l) medium, and the hcp-2 gene expression levels from T6SS were determined in both conditions to analyze if the variability of carbon sources that could happen at the SHIME® model would be involved directly in the T6SS regulation. The stx-2 was also evaluated since it is a key virulence factor in EHEC. The low-glucose level led to an increased expression of the hcp-2 gene both in WT and ΔqseC strains, respectively eightfold and sixfold. In this condition, the absence of the QseC sensor has shown a distinct hcp-2 gene expression when compared to WT levels. Furthermore, the QseC sensor complementation restored the hcp-2 expression similar to the WT strain under the same conditions (Fig. 4A). Seemingly, the glucose levels interfere in the regulation of the hcp-2 gene, and the QseC sensor has a role in its regulation. Notably, the stx-2 gene was 35-fold more expressed in WT and 25-fold in the ΔqseC strains compared to the control group. Also, the QseC sensor complementation returned the stx-2 expression to the levels of the WT strain (Fig. 4B). Moreover, these results suggest that an environmentally scarce source of carbon is sufficient to activate gene expression of a bacterial competition system that is under QseC sensor regulation in EHEC.
Fig. 4.
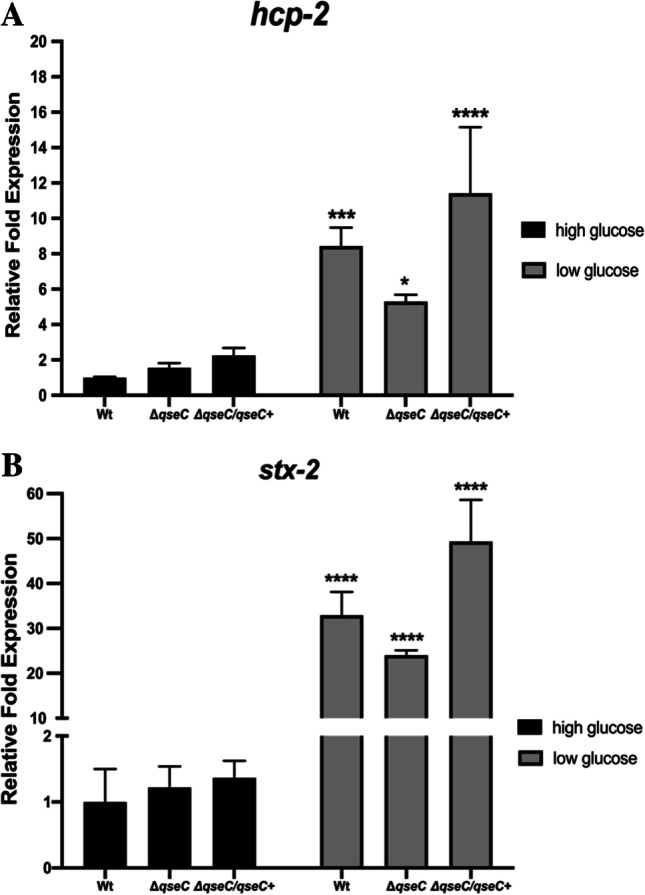
qRT-PCR analysis of the virulence genes expression of the EHEC strains in high and low glucose. The WT, ΔqseC, and ΔqseC/qseC + strains were cultured in low or high glucose in DMEM at 37 °C for 4 h. Then, the qRT-PCR assay was performed with the following genes: A ler and B stx-2. Statistical significance compared to the wild-type strain in high-glucose D-MEM medium. Bars without an asterisk showed no statistically significant difference, p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****) (two-way ANOVA)
B. thetaiotaomicron inhibits the hcp-2 gene expression induction by low-glucose conditions and did not affect EHEC growth
EHEC did not affect Bacteroides genus predominance at SHIME® conditions, and B. thetaiotaomicron did not impair EHEC growth during co-culture; we tested whether hcp-2 gene expression would still be highly expressed under low glucose in co-culture with B. thetaiotaomicron, since T6SS modulation by bacteria is environmental and specie-dependent. Thus, hcp-2 and stx-2 gene expression analyses were performed. There were no significant differences observed in the hcp-2 expression in both WT and ΔqseC strains in low glucose + B. theta when compared to the high glucose + B.theta conditions. These results indicate that the presence of B. thetaiotaomicron was sufficient to decrease the level of hcp-2 expression induced by low glucose. Also, this commensal bacterium may not be a direct competitor to induce T6SS activation in EHEC. When the QseC sensor kinase was restored in the ΔqseC strain, the hcp-2 expression levels were unchanged in high but upregulated in low glucose (Fig. 5A), possibly due to the multiple copy complementation strategy in the ΔqseC/qseC + strain. In both low and high glucose + B. theta conditions, there was no change in stx-2 gene expression (Fig. 5B), which is a different result from that obtained for this gene in the absence of B. thetaiotaomicron in low-glucose conditions (Fig. 4A). These data strongly indicate that B. thetaiotaomicron is not a direct competitor, even when the availability of sugar was decreased, corroborating with the results obtained from SHIME®.
Fig. 5.
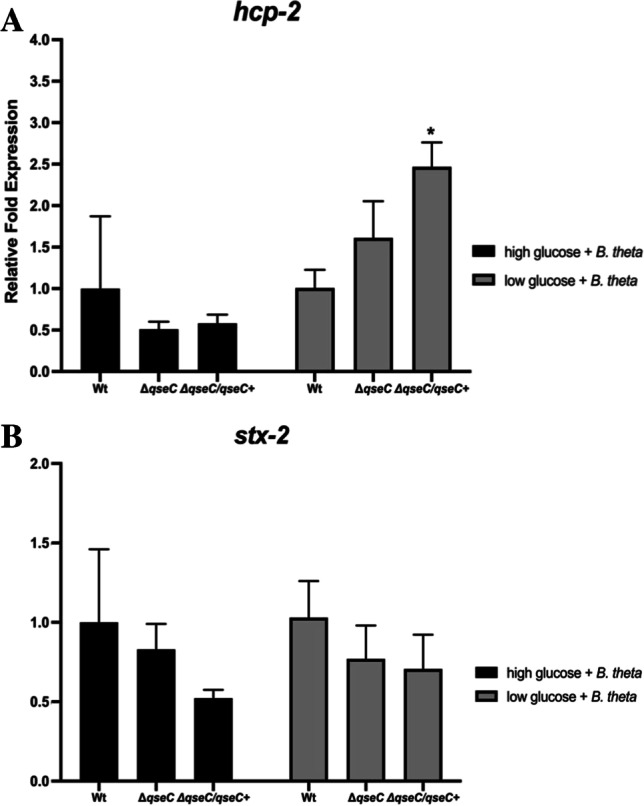
qRT-PCR analysis of virulence gene expression of EHEC strains in low and high glucose in the presence of Bacteroides thetaiotaomicron. The WT, ΔqseC, and ΔqseC/qseC + strains were cultured in low glucose or high-glucose DMEM, anaerobically in the presence of Bacteroides thetaiotaomicron (B. theta) at 37 °C for 4 h. Then, the qRT-PCR assay was performed with the following genes: A ler and B stx-2. Statistical significance compared to the wild-type strain in high glucose + B.theta. Bars without an asterisk showed no statistically significant difference, p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****) (two-way ANOVA)
Different environmental signaling is involved in the hcp-2 gene expression in EHEC, and the QseC sensor has a role in its regulation
Considering the vast cues present in a complex environment such as the gut, several intestinal metabolites such as short fatty acids are known to be key compounds produced by the microbiota that can modulate responses in pathogens [31]. We hypothesized whether fatty acids could have a role in the QseBC signaling in the EHEC T6SS system and would be involved in the SHIME® microbiota modulation by EHEC. Since recent studies have shown that butyrate is a short fatty acid capable of inducing optimal virulence gene expression in EHEC at 20 mM concentration [32], we decided to evaluate the gene expression in media supplemented with this short fatty acid. Expression analyses of hcp-2 and stx-2 genes from strains cultured in the presence of 20 mM butyrate sodium (But) were evaluated. Sodium chloride (NaCl) at 20 mM was used as the osmolarity control for the assays. The gene hcp-2 showed no expression differences in the NaCl treatment nor WT or ΔqseC strains. On the other hand, there was a significant reduction in its expression by But in the ΔqseC strain (Fig. 6A). Corroborating with the hcp-2 results in the NaCl treatment, the stx-2 gene did not show any differences in its expression for both strains; however, there was a slight increase in the expression of this gene in the presence of But for the WT strain (Fig. 6B). Since SHIME® has a complex microbial community under different environmental signals, and the hcp-2 gene was decreased under sugar and short fatty acids signals in ΔqseC strain, the absence of QseC sensor could affect the ability of EHEC to broadly senses environmental signals to activate a system that might be involved its competition with the microbiota and may be involved in the differences observed between the WT and ΔqseC strains at SHIME®.
Fig. 6.
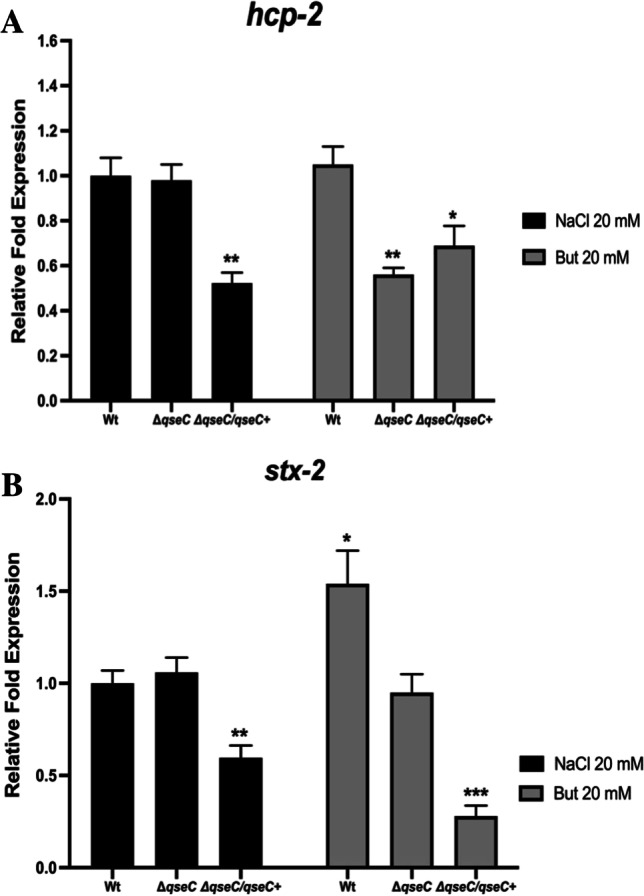
qRT-PCR analysis of the virulence gene expression in the presence of sodium chloride (NaCl) or sodium butyrate (But). The wild-type WT, ΔqseC, and ΔqseC/qseC + strains were cultured in low-glucose DMEM medium at 37 °C in the presence of 20 mM NaCl or But up to O.D of 1. Then, the RT-PCR assay was performed with the following genes: A ler and B stx-2. Statistical significance compared to the wild-type strain in the presence of NaCl. The bars without an asterisk showed no statistically significant difference, p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****) (two-way ANOVA)
L. acidophilus induces considerable EHEC growth inhibition when disputing for the same niche
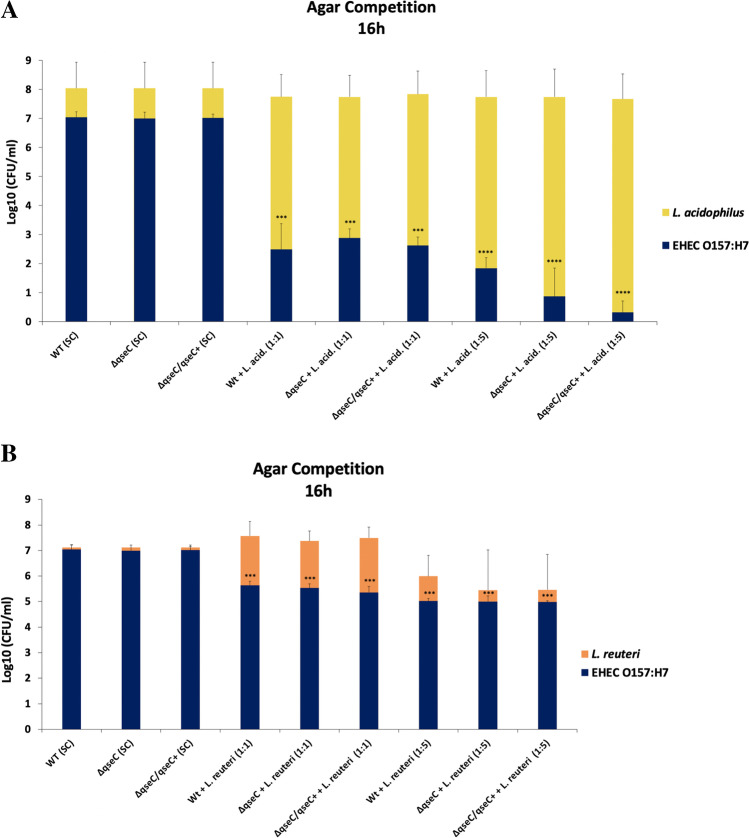
Studies have shown that bacteria such as Vibrio cholerae, Salmonella typhimurium, and Shigella sonnei used their T6SS against commensal bacteria to clear their niche of inhibitory competitors [33–35], and for an efficient T6SS mediate killing, studies have shown that contact between bacteria is necessary [36]. Considering that the hcp-2 gene was downregulated in the ΔqseC strain under different environmental signals, significant differences were observed in the Lactobacillaceae modulation between WT and ΔqseC at the SHIME®. To further investigate whether growth inhibition in L. acidophilus and L. reuteri by EHEC would be different in directing surface interaction, an agar growth competition assay was performed between EHEC strains and Lactobacillaceae species. Individually plated, all EHEC strains without Lactobacillaceae species contact presented constant and lower growth than Lactobacillaceae in the control group. Also, no growth differences were observed between WT and the mutant strains. However, when co-cultured in direct contact with the agar surface in a 1:1 ratio with L. acidophilus, all the EHEC strains significantly inhibited their growth after 16 h of growth. Moreover, when the proportion of L. acidophilus was 1:5, EHEC inhibition was even higher. Thus, the ΔqseC mutant tended to be more affected by the increased ratio of L. acidophilus than the WT strain. Different from what was observed at SHIME®, L. acidophilus did not have its growth significantly affected by EHEC (Fig. 7A). On the other hand, when plated with L. reuteri at 1:1 and 1:5 ratios, all EHEC strains presented similar growth and smaller inhibition (Fig. 7B) compared to demonstrated in co-culture (Fig. 3A). Moreover, the different inhibition observed for EHEC is dependent on the species of Lactobacillaceae present, since L. reuteri did not induce the same inhibition levels in EHEC growth as observed for L. acidophilus. Also, the two species tested did not have their growth impaired by EHEC, which might explain the differences observed at SHIME®.
Fig. 7.
Agar surface competition assay between Lactobacillus acidophilus (A) or Limosilactobacillus reuteri (B) and EHEC strains. The WT, ΔqseC, ΔqseC/qseC + , and L. acidophilus strains or L. reuteri in single culture (SC) were directly added to the agar surface. Then, the strain was mixed in co-culture (WT + L. acid or L. reuteri. and ΔqseC + L. acid or L. reuteri. and ΔqseC/qseC + + L. acid or L. reuteri) at the ratio of 1:1 or 1:5 of EHEC plus L. acid or L. reuteri, respectively, and added to the agar surface. After 16 h, the growth halo from the single-culture and co-culture plates was collected, diluted, plated, and counted. Bars without an asterisk showed no statistically significant difference, p < 0.001 (***), p < 0.0001 (****) (two-way ANOVA)
Discussion
EHEC is an important human gastrointestinal pathogen with great clinical importance associated with food outbreaks, mainly due to undercooked contaminated meat, since EHEC can colonize the gastrointestinal tract of cattle being the main reservoir for these pathogens [1]. To date, the dynamic models to understand how EHEC shapes the microbiota or its behavior in the different compartments of the gastrointestinal tract still demand further studies [16–18]. The human intestinal microbiota is composed of a complex microbial community estimated at 1014 microorganisms that offer a variety of benefits for the host, such as integrity and reshape of the intestinal epithelium, immunity regulatory response, vitamins, energy molecules, resistance against pathogens, and assistance to maintaining the gastrointestinal homeostasis [37]. Since microbiota inhibits several niches and competes for energy sources directly with pathogens, it promotes a process called “resistance to colonization,” which helps the host in the protection against infections [38]. Furthermore, the microbiota is essential for the host’s health, and changes in this microbial community, called dysbiosis, have been associated with susceptibility to infections and various inflammatory diseases [38]. Studies have shown that individuals with Crohn’s disease and ulcerative colitis have a relative increase in Bacteroidetes and a decrease in the abundance of Firmicutes phyla [39]. Also, pathogens that cause gastroenteritis may play a role in the initiation and/or exacerbation of inflammatory bowel diseases [40], so intestinal dysbiosis induced by infectious processes may directly impact the host’s health.
EHEC has a high resistance to low pHs [41], which contributes to its low infectious dose of around 50 to 100 CFUs [1]. Also, EHEC is resistant to bile acids, and studies in a bovine host model showed that the EHEC O157:H7 strain could grow around 15% in bile [42]. Etienne-Mesmin et al. (2011) employed a novel probiotic Saccharomyces cerevisiae strain in the TNO gastrointestinal tract model (TIM, Zeist, Netherlands) multicompartmental system that mimics the human upper gastrointestinal tract, and EHEC was able to grow in the distal portions of the digestive tract model followed by an increase in viability after 1 h in the duodenum [15]. Thus, under favorable conditions, such as neutral pH and dissolution of bile salts in the ileum, there was a significant increase in EHEC growth [15, 43]. Previous studies in the single-step dynamic model of the human colon (ARCOL) and TIM models demonstrated that in general, EHEC is particularly resistant to the gastrointestinal environment [17, 18, 43], and Bacteroidetes and Firmicutes were the most upregulated groups. Therefore, the Bacteroides genus was upregulated when compared to the Lactococcus/Pediococcus/Leuconostoc genus, but there was a different modulation on the microbiota induced among the three donors tested [17].
Herein, our study has shown at SHIME® that even after 1 h of exposure to the pH 2.5, the WT and ΔqseC strains remained viable and presented a minor reduction compared to the initial inoculum (stomach 0 h). During the duodenum reactor passage, when the pancreatic juice started to be added to the system, the WT and ΔqseC strains presented differences in their UFCs recovery, which the WT seemed to replicate; however, the same was not observed for the ΔqseC strain (Fig. 1A). Therefore, the QseC sensor may have a role in activating signals to promote replication after stress conditions, such as acid conditions in the stomach. Moreover, the QseC sensor showed to be involved in microbiota modulation by EHEC and the absence of this sensor favored the Firmicutes phylum (Fig. 1B), suggesting that EHEC may have a considerable impact on the microbiota, and the QseC sensor kinase is directly involved in sensing the intestinal environment to regulate gene expression.
Distinct sugar levels lead to the expression of virulence genes in EHEC [9, 44], and bacteria compete for similar nutrients to survive and colonize the gut [28]. When EHEC was grown solely as monoculture, the low-glucose condition led to an increase in the T6SS gene expression in EHEC, but when B. thetaiotaomicron was added as co-culture under the same conditions, these differences were no longer observed (Figs. 4A and 5A). The analysis with B. thetaiotaomicron demonstrated hcp-2 regulation is also dependent on bacteria in the environment. These results agree with previous studies that indicate B. thetaiotaomicron and EHEC in co-culture did not have their growth impaired, inferring that these two bacteria are not direct competitors [27]. Moreover, the results obtained at SHIME® supported these previous data since the Bacteroides did not show to be affected by the EHEC WT strain in the dynamic model used in the study (Fig. 1C). Besides the SHIME® data, here, we unraveled that the differences in the availability of glucose may lead to an increase in the T6SS hcp-2 gene expression under QseC regulation since this gene was downregulated in the ΔqseC in comparison to WT strain (Fig. 4A and B). During the SHIME® passage, the large amounts of microorganisms promoted an environment with low-carbon sources available for EHEC; in this way, the decrease in hcp-2 in the ΔqseC strain might be involved in the differences in the microbiota modulation observed between the WT and ΔqseC strains. Moreover, these results could suggest that under QseC regulation, in an environment that mimics conditions closer to the intestinal epithelium layer, the T6SS system may be active in EHEC whether to induce cytotoxicity to the host cell epithelium or to inhibit possible competitors of this carbon source. Lastly, the stx-2 gene had an overexpression in the low-glucose assay and was decreased in the ΔqseC strain, which corroborated with the differences in stx-2 expression between the WT and ΔqseC observed at SHIME® infection.
The Lactobacillaceae was the bacterial group analyzed most affected by the absence of the QseC sensor (Fig. 1C). When investigating whether it may be a direct competitor with EHEC, our results suggest that the differences in the Lactobacillaceae members at SHIME® might not be due to direct competition between EHEC and Lactobacillaceae. EHEC growth was inhibited during Lactobacillaceae strains co-culture experiments (Figs. 3A, B and 7A, B); however, during SHIME® microbiota analyses, the absence of the QseC sensor kinase led to a significant increase of the Lactobacillaceae. Studies have demonstrated that the Lacticaseibacillus casei (Lactobacillus casei) LC2W inhibited the colonization of EHEC in mice [45]. Similarly, the administration of L. reuteri before and during infection by EHEC in germ-free mice resulted in the improvement of the disease symptoms and increased protection from EHEC infection in mice [46]. Thus, Lactobacillaceae species can produce a broad range of bacteriocins such as helveticin and lactocillin against different bacteria [47], and some gram-positive bacteria species, such as Bacillus and Listeria, possess the wall-associated protein A (WapA) that seems to be contact-dependent to promote bacterial growth inhibition [48, 49]. Also, previous studies have demonstrated that L. acidophilus through the production of bioactive molecules could impair EHEC virulence [50], and the high production of linoleic acid by L. casei limited the growth, survival, and virulence of EHEC and Salmonella typhimurium [51], which corroborates to our data here. Herein, the results suggest that L. acidophilus may have a competition niche system that is contact-dependent to inhibit EHEC growth, but the QseC sensor does not seem to help EHEC to survive in this condition. Furthermore, the differences observed for Lactobacillaceae between the strains during the in vitro co-culture and SHIME® analysis may be due to the dynamic model; there is a pool of Lactobacillaceae species that is possibly affected by EHEC, and it was not evaluated under co-culture conditions here performed. Also, the absence of the QseC sensor could lead to ineffective competition between the ΔqseC strain and other members of the microbiota that may help the Lactobacillaceae family at SHIME®. Nevertheless, the significant large inhibition promoted by L. acidophilus and the modulations observed at SHIME® opens a perspective that members of the Lactobacillaceae genus might be an important competitor for EHEC. Additionally, short fatty acids seem to be involved in the T6SS system regulation promoted by the QseC sensor (Fig. 6A and B), indicating the broad regulatory signals that may be under the control of this sensor and its role in EHEC pathogenesis.
Conclusions
The QseC sensor kinase modulates the gut microbiota within the conditions here tested, and its regulation under distinct glucose and butyrate concentrations is crucial to T6SS gene-encoding factors in EHEC. Therefore, this study brings new insights into the QseC sensor role that impacts the activation of LEE virulence factors and the direct competition of EHEC with the microbiota, contributing to a broad response to different metabolites and signals in the intestinal environment by EHEC. Our gut microbiome results have shown that the QseC sensor inhibition has an important impact on pathogen virulence, supporting the idea that QseC-blocking could be an interesting target during the gut microbiota competition for novel therapies. Further studies will help to understand how the microbiota modulation and intestinal metabolites under QseC sensor control contribute to EHEC colonization, as well as the benefits of an anti-virulence approach against pathogens as an alternative conventional therapeutic approach.
Acknowledgements
We thank all members of the Biological Sciences Department at the Faculty of Pharmaceutical Sciences, São Paulo State University.
Author contribution
K.M. and C.G.M. designed the study. KM performed the research. K.M. and C.G.M. performed the data analysis. M.K.S., S.V., and K.M. performed and designed the SHIME assay. K.M. wrote the manuscript. C.G.M. supervised the research.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), grant number 2018/22042–0 and 2019/03049–7, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), grant number 441884/2014–8 and financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Data availability
All data and materials are available under request.
Code availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors consent for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Responsible Editor: Luis Nero
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kaper JB, Nataro JP, Mobley HL. Pathogenic escherichia coli. Nat Rev Microbiol. 2004;2(2):123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet. 1983;322(8362):1299–1300. doi: 10.1016/S0140-6736(83)91167-4. [DOI] [PubMed] [Google Scholar]
- 4.Kimmitt PT, Harwood CR, Barer MR. Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis. 2000;6(5):458. doi: 10.3201/eid0605.000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng W, Li Y, Vallance BA, Finlay BB. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect Immun. 2001;69(10):6323–6335. doi: 10.1128/IAI.69.10.6323-6335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nataro JP, Kaper JB. Diarrheagenic escherichia coli. Clin Microbiol Rev. 1998;11(1):142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatt S, Romeo T, Kalman D. Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol. 2011;19(5):217–224. doi: 10.1016/j.tim.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50(1):727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 9.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. MBio. 2012;3(5):e00280–e312. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria–host communication: the language of hormones. Proc Natl Acad Sci. 2003;100(15):8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sircili MP, Walters M, Trabulsi LR, Sperandio V. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect Immun. 2004;72(4):2329–2337. doi: 10.1128/IAI.72.4.2329-2337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reading NC, Torres AG, Kendall MM, Hughes DT, Yamamoto K, Sperandio V. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol. 2007;189(6):2468–2476. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walters M, Sperandio V. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun. 2006;74(10):5445–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Njoroge J, Sperandio V. Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases. QseC and QseE Infect Immun. 2012;80(2):688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jubelin G, Desvaux M, Schüller S, Etienne-Mesmin L, Muniesa M, Blanquet-Diot S. Modulation of enterohaemorrhagic Escherichia coli survival and virulence in the human gastrointestinal tract. Microorganisms. 2018;6(4):115. doi: 10.3390/microorganisms6040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roussel C, Cordonnier C, Galia W, et al. Increased EHEC survival and virulence gene expression indicate an enhanced pathogenicity upon simulated pediatric gastrointestinal conditions. Pediatr Res. 2016;80(5):734–743. doi: 10.1038/pr.2016.144. [DOI] [PubMed] [Google Scholar]
- 17.Thevenot J, Cordonnier C, Rougeron A, et al. Enterohemorrhagic Escherichia coli infection has donor-dependent effect on human gut microbiota and may be antagonized by probiotic yeast during interaction with Peyer’s patches. Appl Microbiol Biotechnol. 2015;99(21):9097–9110. doi: 10.1007/s00253-015-6704-0. [DOI] [PubMed] [Google Scholar]
- 18.Thévenot J, Etienne-Mesmin L, Denis S, Chalancon S, Alric M, Livrelli V, Blanquet-Diot S. Enterohemorrhagic Escherichia coli O157: H7 survival in an in vitro model of the human large intestine and interactions with probiotic yeasts and resident microbiota. Appl Environ Microbiol. 2013;79(3):1058–1064. doi: 10.1128/AEM.03303-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van de Wiele T, Van den Abbeele P, Ossieur W, Possemiers S, & Marzorati M (2015) The simulator of the human intestinal microbial ecosystem (SHIME®). The Impact of Food Bioactives on Health: in vitro and ex vivo models, E-book, chapter 27:305–317
- 20.Collins JW, Keeney KM, Crepin VF, Rathinam VA, Fitzgerald KA, Finlay BB, Frankel G. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol. 2014;12(9):612–623. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- 21.Molly K, Woestyne MV, Smet ID, Verstraete W. Validation of the simulator of the human intestinal microbial ecosystem (SHIME) reactor using microorganism-associated activities. Microb Ecol Health Dis. 1994;7(4):191–200. [Google Scholar]
- 22.Bianchi F, Larsen N, de Mello Tieghi T, et al. Modulation of gut microbiota from obese individuals by in vitro fermentation of citrus pectin in combination with Bifidobacterium longum BB-46. Appl Microbiol Biotechnol. 2018;102(20):8827–8840. doi: 10.1007/s00253-018-9234-8. [DOI] [PubMed] [Google Scholar]
- 23.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC) PLoS pathogens. 2009;5(8):e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shigeno T, Akamatsu T, Fujimori K, Nakatsuji Y, Nagata A. The clinical significance of colonoscopy in hemorrhagic colitis due to enterohemorrhagic Escherichia coli O157: H7 infection. Endosc. 2002;34(04):311–314. doi: 10.1055/s-2002-23644. [DOI] [PubMed] [Google Scholar]
- 25.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp in ulcerative colitis. J Med Microbiol. 2006;55(5):617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 26.Kaakoush NO, Day AS, Huinao KD, Leach ST, Lemberg DA, Dowd SE, Mitchell HM. Microbial dysbiosis in pediatric patients with Crohn’s disease. J Clin Microbiol. 2012;50(10):3258–3266. doi: 10.1128/JCM.01396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16(6):759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raffatellu M. Learning from bacterial competition in the host to develop antimicrobials. Nat Med. 2018;24(8):1097–1103. doi: 10.1038/s41591-018-0145-0. [DOI] [PubMed] [Google Scholar]
- 29.Cascales E. The type VI secretion toolkit. EMBO Rep. 2008;9(8):735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan B, Zhang Q, Ni J, et al. Type VI secretion system contributes to Enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS) PLoS Pathog. 2017;13(3):e1006246. doi: 10.1371/journal.ppat.1006246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci. 1995;92(17):7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiol. 2009;155(2):521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 33.Sana TG, Flaugnatti N, Lugo KA, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci. 2016;113(34):E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao W, Caro F, Robins W, Mekalanos JJ. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Sci. 2018;359(6372):210–213. doi: 10.1126/science.aap8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol. 2008;47(5):367–373. doi: 10.1111/j.1472-765X.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- 36.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12(2):137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Lopez A, Behnsen J, Nuccio SP, Raffatellu M. Mucosal immunity to pathogenic intestinal bacteria. Nat Rev Immunol. 2016;16(3):135–148. doi: 10.1038/nri.2015.17. [DOI] [PubMed] [Google Scholar]
- 39.Rapozo DC, Bernardazzi C, de Souza HSP. Diet and microbiota in inflammatory bowel disease: The gut in disharmony. World J Gastroenterol. 2017;23(12):2124. doi: 10.3748/wjg.v23.i12.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodríguez LAG, Ruigómez A, Panés J. Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterol. 2006;130(6):1588–1594. doi: 10.1053/j.gastro.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Benjamin MM, Datta AR. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61(4):1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoffregen WC, Pohlenz JF, Dean-Nystrom EA. Escherichia coli O157: H7 in the gallbladders of experimentally infected calves. J Vet Diagn Invest. 2004;16(1):79–83. doi: 10.1177/104063870401600114. [DOI] [PubMed] [Google Scholar]
- 43.Etienne-Mesmin L, Livrelli V, Privat M, Denis S, Cardot JM, Alric M, Blanquet-Diot S. Effect of a new probiotic Saccharomyces cerevisiae strain on survival of Escherichia coli O157: H7 in a dynamic gastrointestinal model. Appl Environ Microbiol. 2011;77(3):1127–1131. doi: 10.1128/AEM.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Wu Y, Wang Y, et al. Antioxidant properties of probiotic bacteria. Nutrients. 2017;9(5):521. doi: 10.3390/nu9050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang G, Zhang Y, Song X, Xia Y, Lai PFH, Ai L. Lactobacillus casei LC2W can inhibit the colonization of Escherichia coli O157: H7 in vivo and reduce the severity of colitis. Food Funct. 2019;10(9):5843–5852. doi: 10.1039/C9FO01390C. [DOI] [PubMed] [Google Scholar]
- 46.Eaton KA, Honkala A, Auchtung TA, Britton RA. Probiotic Lactobacillus reuteri ameliorates disease due to enterohemorrhagic Escherichia coli in germfree mice. Infect Immun. 2011;79(1):185–191. doi: 10.1128/IAI.00880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García-Bayona L, Comstock LE. Bacterial antagonism in host-associated microbial communities. Sci. 2018;361(6408):eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 48.Willett JL, Ruhe ZC, Goulding CW, Low DA, Hayes CS. Contact-dependent growth inhibition (CDI) and CdiB/CdiA two-partner secretion proteins. J Mol Biol. 2015;427(23):3754–3765. doi: 10.1016/j.jmb.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koskiniemi S, Lamoureux JG, Nikolakakis KC, de Roodenbeke CTK, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci. 2013;110(17):7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Medellin-Pena MJ, Griffiths MW. Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157: H7 colonization. Appl Environ Microbiol. 2009;75(4):1165–1172. doi: 10.1128/AEM.01651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng M, Tabashsum Z, Patel P, Bernhardt C, & Biswas D (2018) Linoleic acids overproducing Lactobacillus casei limits growth, survival, and virulence of Salmonella Typhimurium and enterohaemorrhagic Escherichia coli. Front. Microbiol 9:2663 [DOI] [PMC free article] [PubMed]
- 52.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli 0157: H7 infections: a broad clinical spectrum. Ann Intern Med. 1988;109(9):705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 53.Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E coli. Mol Microbiol. 2002;43(3):809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 54.Rasko DA, Moreira CG, Li DR, et al. Targeting QseC signaling and virulence for antibiotic development. Sci. 2008;321(5892):1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the prevotella-to-bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl Environ Microbiol. 2014;80(3):1142–1149. doi: 10.1128/AEM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Gregoris TB, Aldred N, Clare AS, Burgess JG. Improvement of phylum-and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods. 2011;86(3):351–356. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 57.Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71(7):4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97(6):1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials are available under request.
Not applicable.