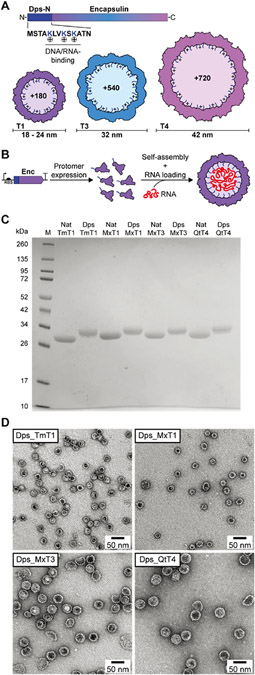
Figure 1.
Design and initial characterization of the encapsulin-based in vivo RNA encapsulation system. (A) Schematic of the engineered encapsulin protomer sequence with the N-terminally fused nucleic acid-binding peptide Dps-N. The three different sizes (T1, T3, and T4) of engineered encapsulins (Dps_Encs) created in this study are shown, highlighting the additional luminal positive charges introduced by Dps-N. T: triangulation number (T-number). (B) Schematic outlining the principle of in vivo nucleic acid encapsulation by Dps_Encs. Enc: encapsulin. (C) SDS-PAGE analysis and comparison of purified native (Nat) and engineered (Dps) encapsulins used in this study. Encapsulins are labeled by their organism of origin and T-number. Tm: Thermotoga maritima, Mx: Myxococcus xanthus, Qt: Quasibacillus thermotolerans. (D) Negative-stain TEM micrographs of all four purified Dps_Encs used in this study.