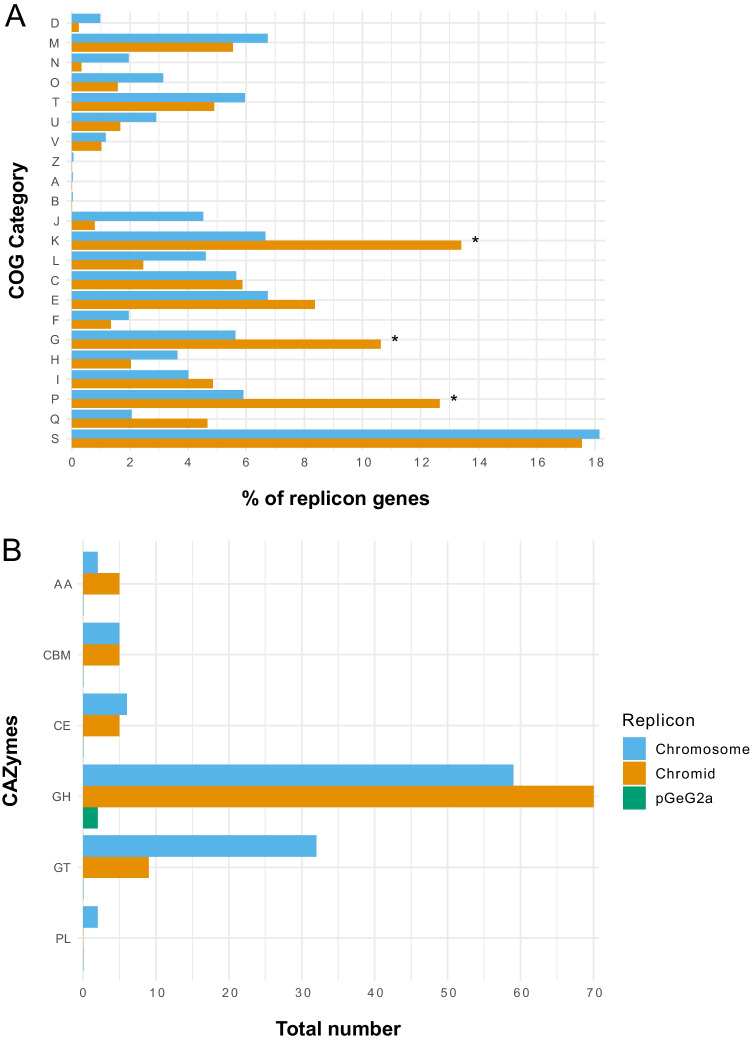
Fig. 6.
COG functional categories (A) and carbohydrate-active enzymes (CAZymes) (B) predicted in each replicon of strain GeG2T genome. COG categories with significantly different proportions as shown by the two-proportions Z-test are indicated with *. D: cell cycle control, cell division, chromosome partitioning; M: cell wall/membrane/envelop biogenesis; N: cell motility; O: post-translational modification, protein turnover, chaperone functions; T: signal transduction mechanisms; U: intracellular trafficking, secretion, and vesicular transport; V: defense mechanisms; Z: cytoskeleton; A: RNA processing and modification; B: chromatin structure and dynamics; J: translation, ribosomal structure, and biogenesis; K: transcription; L: replication and repair; C: energy production and conversion; E: amino acid metabolism and transport; F: nucleotide metabolism and transport; G: carbohydrate metabolism and transport; H: coenzyme metabolism and transport; I: lipid metabolism and transport; P: inorganic ion transport and metabolism; Q: secondary metabolites biosynthesis, transport, and catabolism; S: function unknown. GT: glycosyl transferases; GH: glycosyl hydrolases; CE: carbohydrate esterases; CBM: carbohydrate-binding module; AA: enzymes for the auxiliary activities; PL: polysaccharide lyases