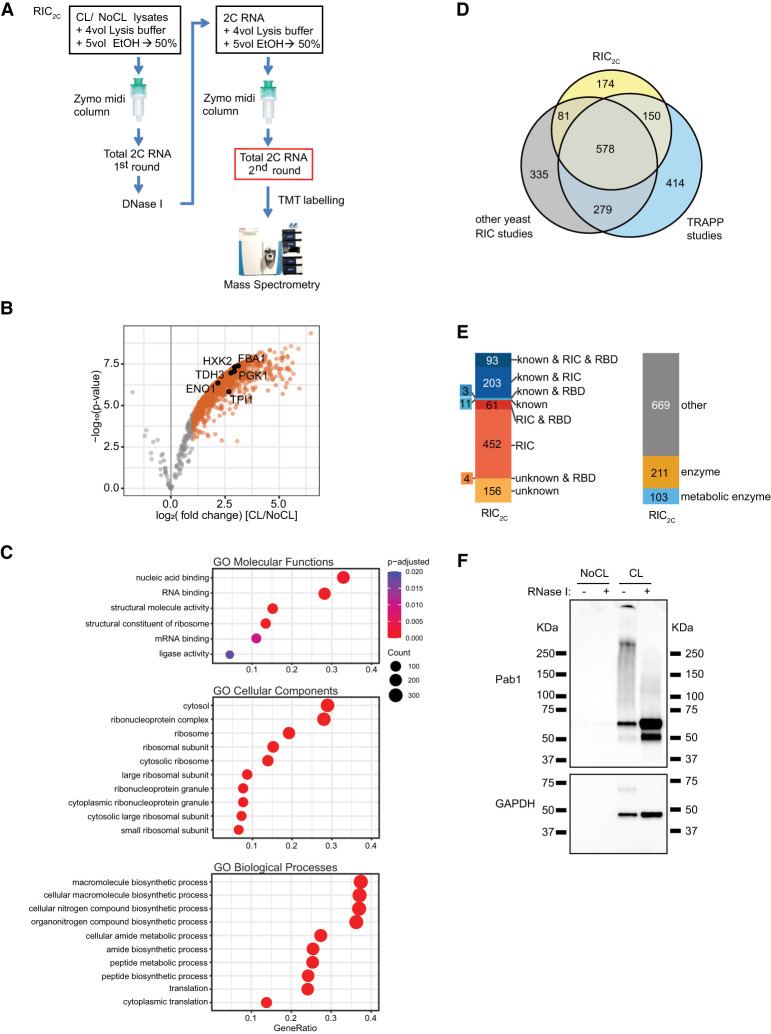
FIGURE 1.
2C total RNA interactome capture (RIC2C) identified 983 RBPs in yeast. (A) Schematic representation of RIC2C. UV-cross-linked and nonirradiated negative controls were subjected to a first round of 2C. Any residual DNA in the eluates was digested by DNase I, and the RNA and RNA–protein adducts were repurified by a second 2C extraction. Eluates from the second round were RNase I–treated and proteins were subjected to TMT labeling and mass spectrometry analysis. (B) Volcano plot displaying log2 fold change of protein abundance versus −log10 P-value after RIC2C of CL and NoCL samples. Gray dots represent proteins displaying no statistically significant difference. Orange dots represent proteins statistically enriched (logFC ≥ 1 and P-value ≤ 0.05) in CL over NoCL samples. Glycolytic enzymes statistically enriched in the CL fraction are highlighted with black circles. (C) Top 10 significantly (Padjusted < 0.05) enriched GO molecular functions, cellular components and biological processes terms of RBPs identified by RIC2C compared to the identified background. (D) Venn diagram showing the overlap between the RBPs detected by RIC2C, TRAPP (Shchepachev et al. 2019) and a compendium of other RNA interactome capture experiments in yeast (Scherrer et al. 2010; Tsvetanova et al. 2010; Mitchell et al. 2013; Ray et al. 2013; Kramer et al. 2014; Beckmann et al. 2015; Matia-Gonzalez et al. 2015; Brannan et al. 2016; Shchepachev et al. 2019). (E) Analysis of the RBPs detected after RIC2C in yeast. RBPs were categorized according to experimental evidence described in literature (“known”), their detection on RIC experiments (RIC) or content of RNA binding domains (RBD). Novel RBPs detected by RIC2C unrelated to previous experimental evidence and not detected on any RIC experiment were categorized as “unknown.” (F) Validation of two RBPs by 2C-western blot. A total of 10 µg of 2C RNA from CL and NoCL samples were treated or not with RNase I, separated by SDS-PAGE, blotted to a nitrocellulose membrane and probed against Pab1 and GAPDH antibodies.