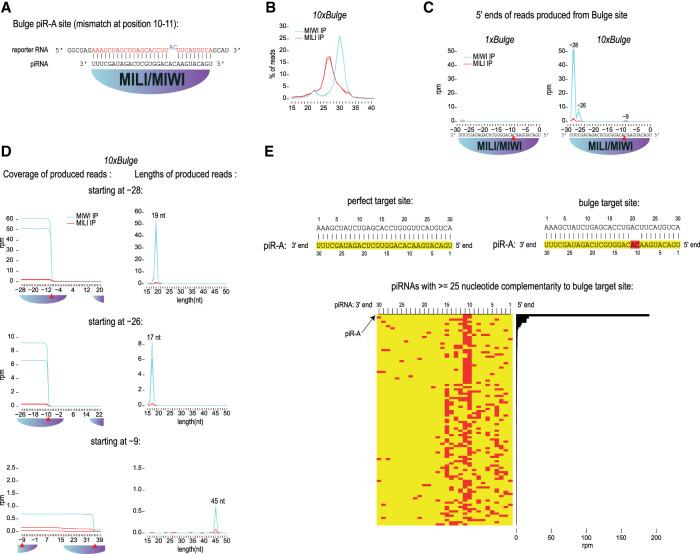
FIGURE 2.
RNA targets with cleavage-site mismatches are sliced by PIWI proteins in vivo. (A) The bulge piR-A sites were designed to contain noncomplementary bases to piR-A nucleotides at positions 10 and 11. (B) Length profiles of immunoprecipitated small RNAs from 10xBulge mouse. (C) The position (=distance from 5′ nt of targeting piR-A) and abundance of 5′ ends of reads produced from reporter target sites. In the case of 10xBulge, the metaplot summarizes the read counts from all ten sites. Red triangle refers to expected position of MILI/MIWI cleavage guided by piR-A. (D) Coverage of reads is shown separately for RNAs with their 5′ end starting at a specific distance from the first piR-A target nucleotide (=0 distance). These RNAs are of specific lengths, with the 3′ ends always created by MILI/MIWI slicing at a subsequent site. (E) Complementary piRNAs potentially targeting the bulge site. The heatmap depicts position of mismatches (in red) between the targeting piRNA and the target site sequence, while the barplot shows the abundance of such piRNAs.