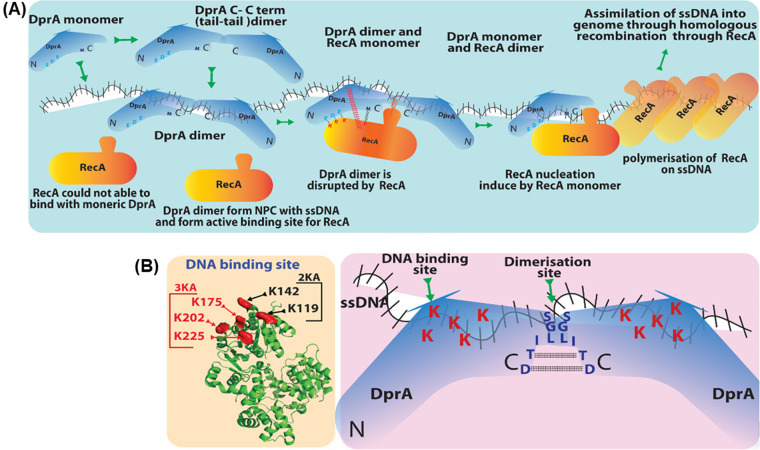
FIG 7.
(A) DprA-mediated loading of RecA on ssDNA. DprASp dimer (C-C term) forms a nucleoprotein complex with ssDNA. DprASp dimer bound to ssDNA offers the best suitable platform for RecA nucleation. After ssDNA binding of DprASp dimer conformation change takes place, and sites of DprASp-RecA interaction open up (EDE triad of DprASp interacts with RRK triad of RecA). Once DprASp-RecA interaction is established, DprASp is converted to a monomer. This change allows RecA loading on ssDNA, followed by RecA nucleation, continued polymerization, and DprASp leaving the ssDNA. (B) Residues important for S. pneumoniae DprA protein DNA binding and dimerization. Five lysine (K119/K144/K175/K202/K225) residues on the basic face of DprASp are essential for DNA binding activity. DNA binding activity is completely abolished when lysine is mutated to alanine, whereas the partial patch mutation 3KA and 2KA produces a similar effect equal to that of R115A single mutant. This basic patch is localized opposite the RecA binding site.