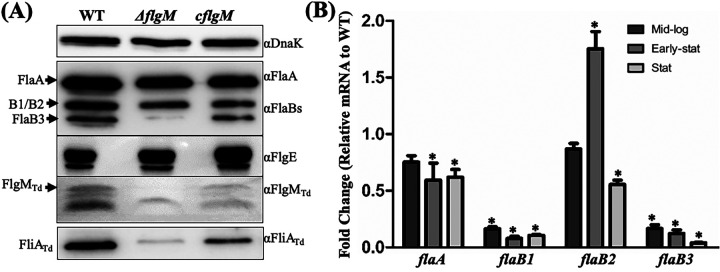
FIG 6.
Deletion of flgMTd impairs T. denticola flagellar filament gene expression. (A) Immunoblotting analysis. Equivalent amounts of whole-cell lysates of the wild-type (WT), ΔflgM, and cflgM strains were analyzed by SDS-PAGE and then probed with specific antibodies against the following proteins: DnaK (loading control), FlaA, FlaBs, FlgE, FlgMTd, and FliATd. (B) qRT-PCR analysis. For this experiment, the levels of four flagellar filament genes (flaA, flaB1, flaB2, and flaB3) were measured by qRT-PCR as previously described (41, 48). The dnaK gene transcript was used as an internal control to normalize the readouts of qRT-PCR. The results are expressed as the level of individual gene transcripts in the ΔflgM mutant relative to that of the wild type at three different growth phases, including the mid-logarithmic (Mid-log), early stationary (Early-stat), and stationary (Stat) phases. Asterisks indicate that the difference between the wild-type and ΔflgM strains was statistically significant at a P value of <0.01 (one-way ANOVA followed by Tukey’s multiple-comparison test).